Sequence Alignment Topics Introduction Exact Algorithm Alignment Models
![The IDEA s[1…n] t[1…m] To align s[1. . . i] with t[1…j] we have The IDEA s[1…n] t[1…m] To align s[1. . . i] with t[1…j] we have](https://slidetodoc.com/presentation_image_h/b74237f36a7493dbf1200fdcaf2f0c2a/image-7.jpg)


- Slides: 28

Sequence Alignment Topics: • Introduction • Exact Algorithm • Alignment Models • Bio. Perl functions

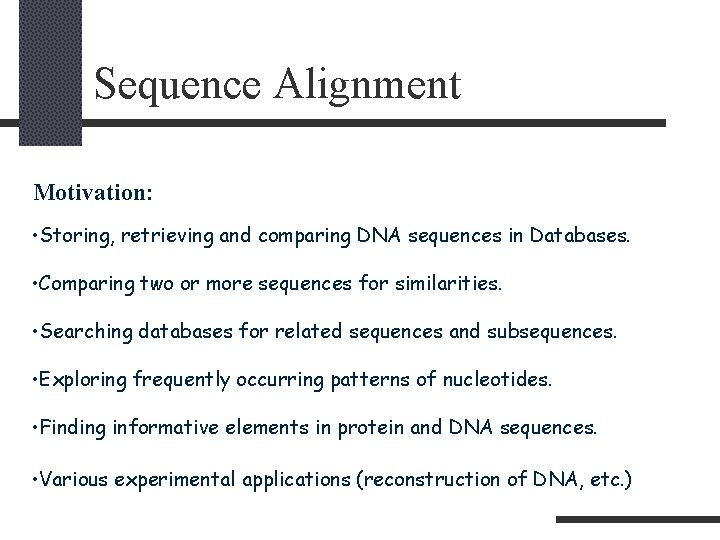
Sequence Alignment Motivation: • Storing, retrieving and comparing DNA sequences in Databases. • Comparing two or more sequences for similarities. • Searching databases for related sequences and subsequences. • Exploring frequently occurring patterns of nucleotides. • Finding informative elements in protein and DNA sequences. • Various experimental applications (reconstruction of DNA, etc. )

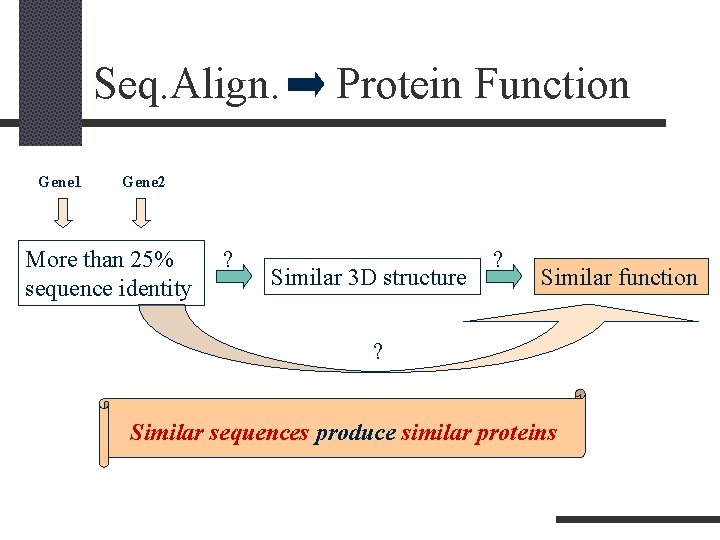
Seq. Align. Gene 1 Protein Function Gene 2 More than 25% sequence identity ? Similar 3 D structure ? Similar function ? Similar sequences produce similar proteins

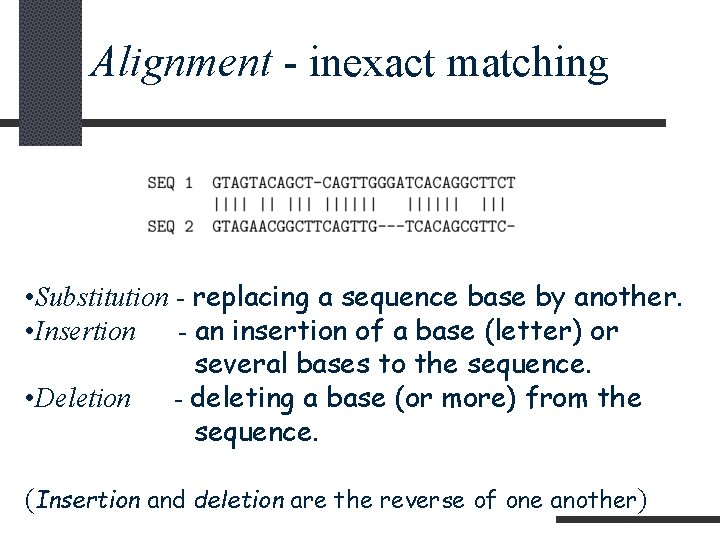
Alignment - inexact matching • Substitution - replacing a sequence base by another. • Insertion - an insertion of a base (letter) or several bases to the sequence. • Deletion - deleting a base (or more) from the sequence. (Insertion and deletion are the reverse of one another)

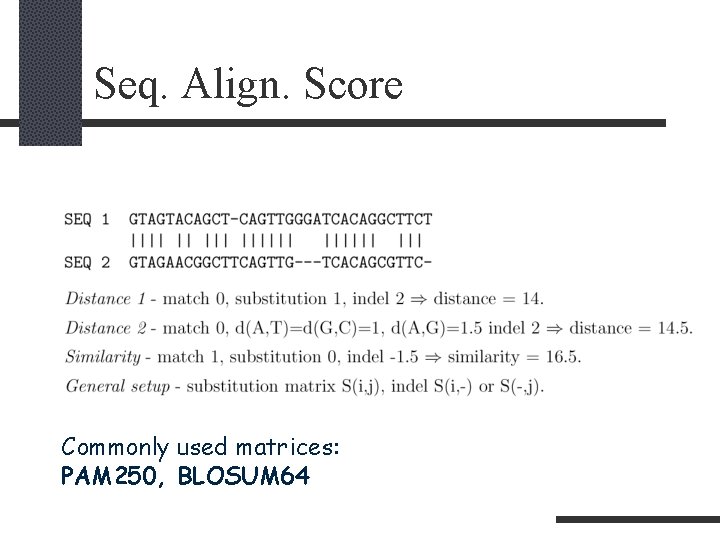
Seq. Align. Score Commonly used matrices: PAM 250, BLOSUM 64

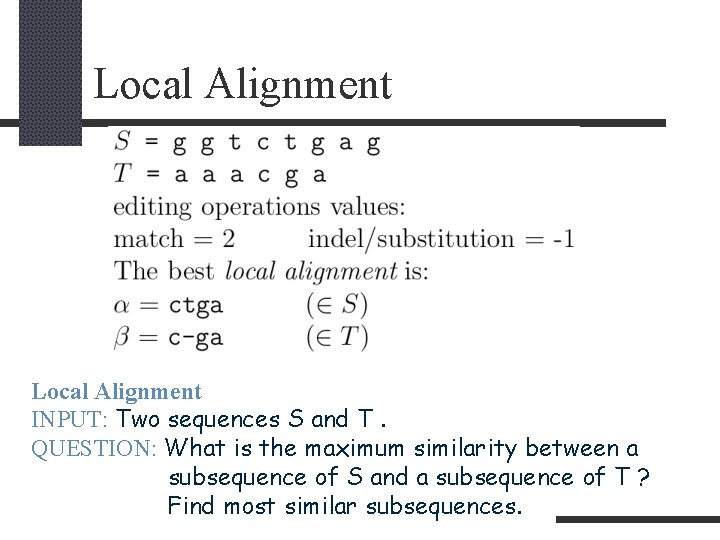
Local Alignment INPUT: Two sequences S and T. QUESTION: What is the maximum similarity between a subsequence of S and a subsequence of T ? Find most similar subsequences.
![The IDEA s1n t1m To align s1 i with t1j we have The IDEA s[1…n] t[1…m] To align s[1. . . i] with t[1…j] we have](https://slidetodoc.com/presentation_image_h/b74237f36a7493dbf1200fdcaf2f0c2a/image-7.jpg)
The IDEA s[1…n] t[1…m] To align s[1. . . i] with t[1…j] we have three choices: * align s[1…i-1] with t[1…j-1] and match s[i] with t[j] s[1…i-1] i t[1…j-1] j * align s[1…i] with t[1…j-1] and match a space with t[j] s[1… i ] t[1…j-1] j * align s[1…i-1] with t[1…j] and match s[i] with a space s[1…i-1] i t[1… j ] -

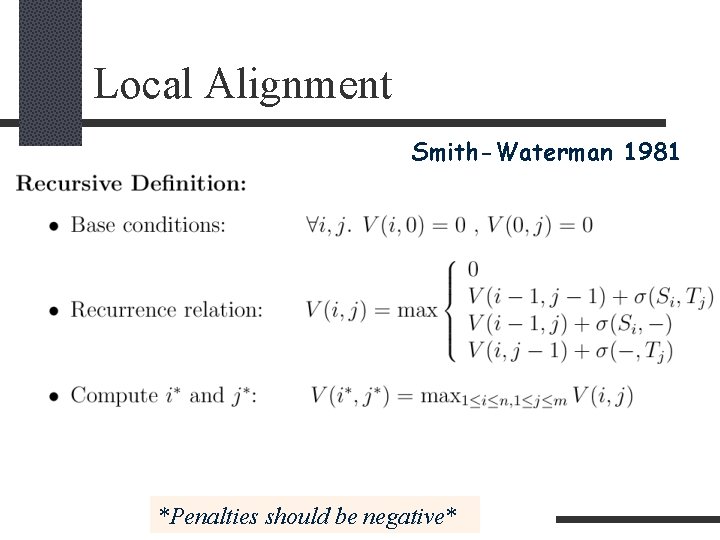
Local Alignment Smith-Waterman 1981 *Penalties should be negative*

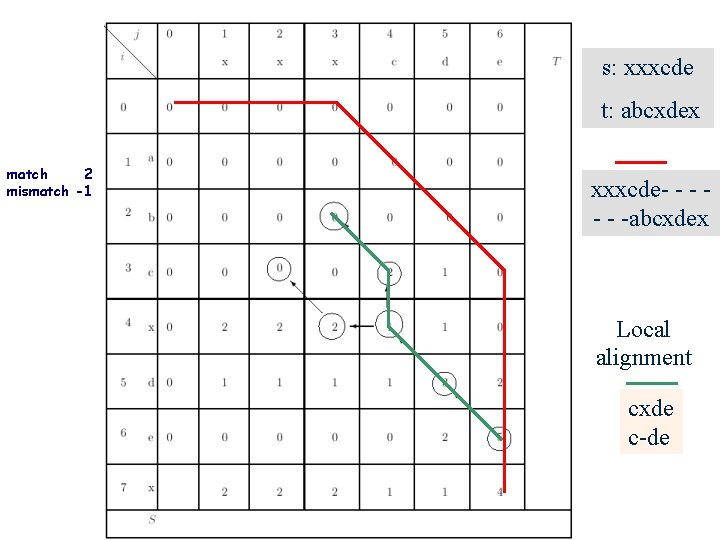
s: xxxcde t: abcxdex match 2 mismatch -1 xxxcde- - - -abcxdex Local alignment cxde c-de

Sequence Alignment Complexity: Time O(n*m) Space O(n*m) (exist algorithm with O(min(n, m)) )

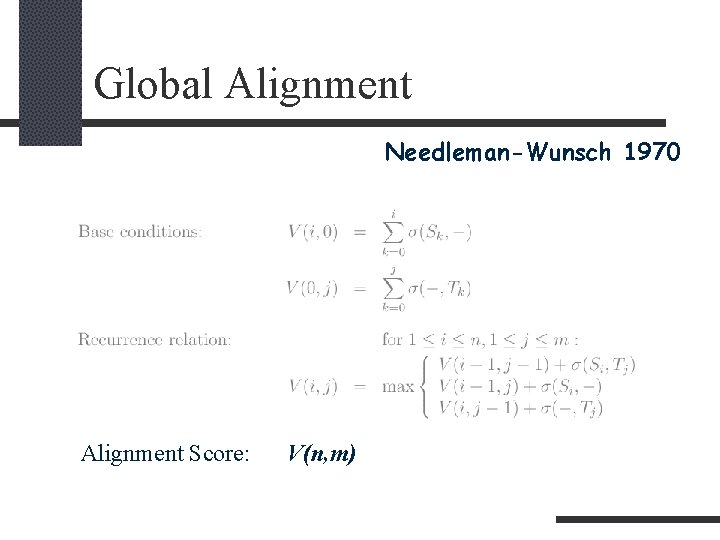
Global Alignment INPUT: Two sequences S and T of roughly the same length. QUESTION: What is the maximum similarity between them? Find one of the best alignments.

Global Alignment Needleman-Wunsch 1970 Alignment Score: V(n, m)

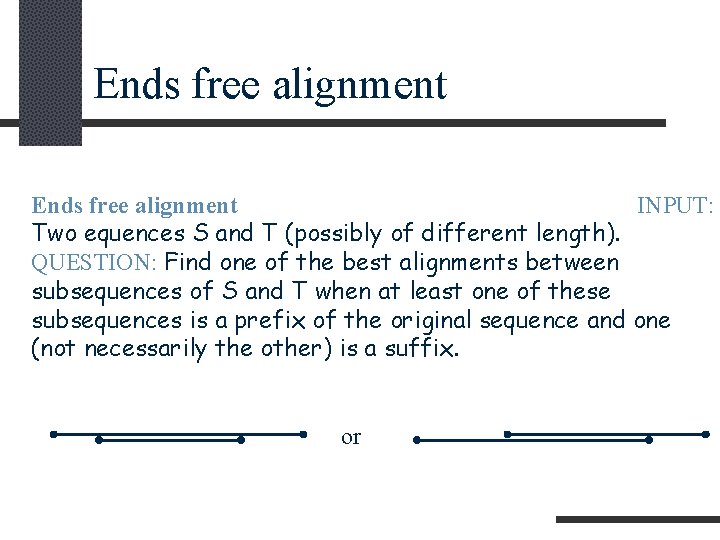
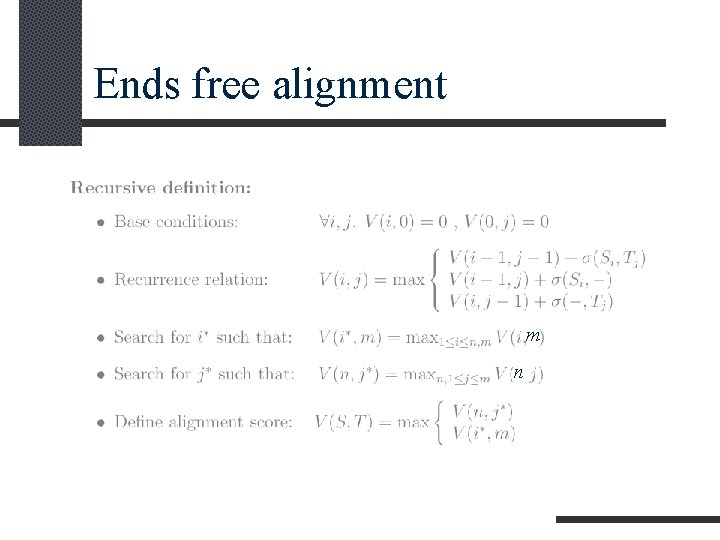
Ends free alignment INPUT: Two equences S and T (possibly of different length). QUESTION: Find one of the best alignments between subsequences of S and T when at least one of these subsequences is a prefix of the original sequence and one (not necessarily the other) is a suffix. or

Ends free alignment m n

Gap Alignment Definition: A gap is the maximal contiguous run of spaces in a single sequence within a given alignment. The length of a gap is the number of indel operations on it. A gap penalty function is a function that measure the cost of a gap as a (nonlinear) function of its length. Gap penalty INPUT: Two sequences S and T (possibly of different length). QUESTION: Find one of the best alignments between the two sequences using the gap penalty function. Affine Gap: Wtotal = Wg + q. Ws Wg – weight to open the gap Ws – weight to extend the gap

Bio. Perl “Bioperl is a collection of perl modules that facilitate the development of perl scripts for bio-informatics applications. ” Bioperl is open source software that is still under active development. www. bioperl. org Tutorial Documentation

Bio. Perl • Accessing sequence data from local and remote databases • Transforming formats of database/ file records • Manipulating individual sequences • Searching for "similar" sequences • Creating and manipulating sequence alignments • Searching for genes and other structures on genomic DNA • Developing machine readable sequence annotations

Bio. Perl library at TAU Bio. Perl is NOT yet installed globally on CS network. In each script you should add the following two lines: use lib "/a/netapp/vol 0/home/silly 6/mol/lib/Bio. Perl/lib"; use lib "/a/netapp/vol 0/home/silly 6/mol/lib/Bio. Perl/lib/i 686 -linux";

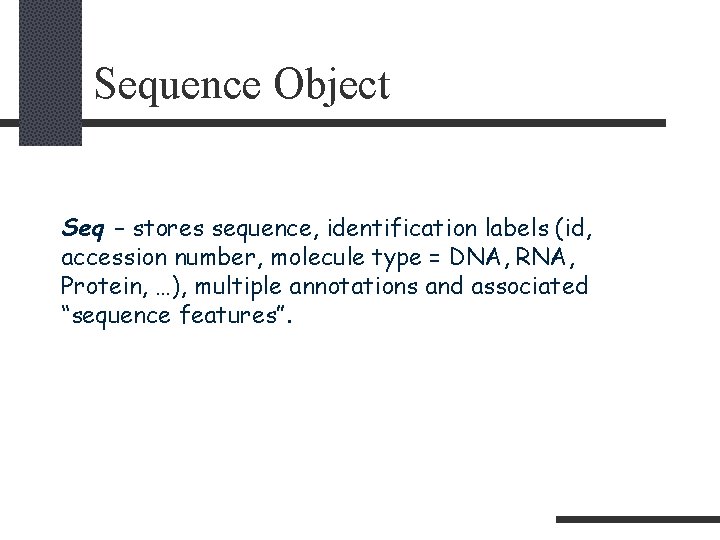
Sequence Object Seq – stores sequence, identification labels (id, accession number, molecule type = DNA, RNA, Protein, …), multiple annotations and associated “sequence features”.

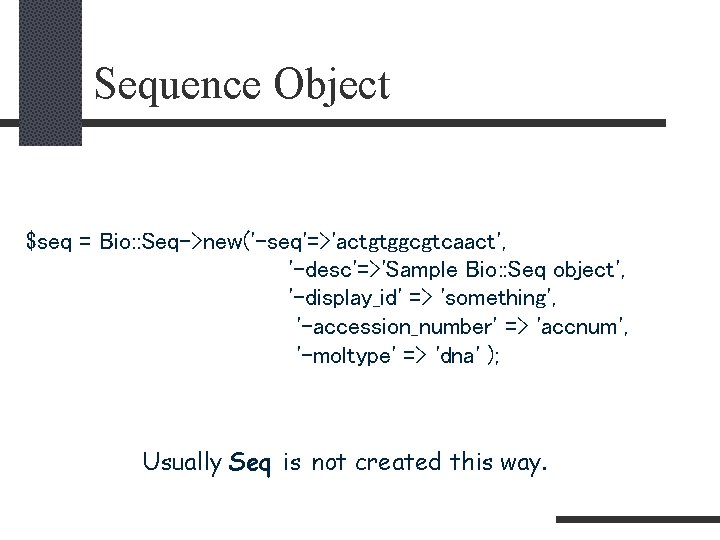
Sequence Object $seq = Bio: : Seq->new('-seq'=>'actgtggcgtcaact', '-desc'=>'Sample Bio: : Seq object', '-display_id' => 'something', '-accession_number' => 'accnum', '-moltype' => 'dna' ); Usually Seq is not created this way.

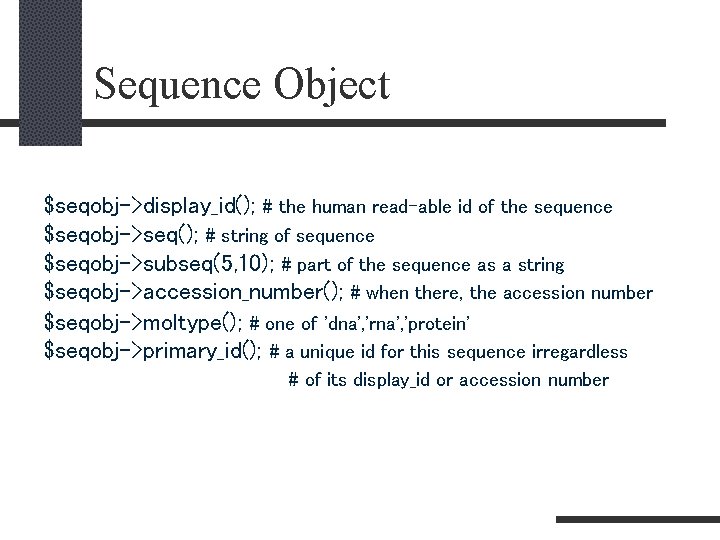
Sequence Object $seqobj->display_id(); # the human read-able id of the sequence $seqobj->seq(); # string of sequence $seqobj->subseq(5, 10); # part of the sequence as a string $seqobj->accession_number(); # when there, the accession number $seqobj->moltype(); # one of 'dna', 'rna', 'protein' $seqobj->primary_id(); # a unique id for this sequence irregardless # of its display_id or accession number

Accessing Data Base Databases: genbank, genpept, swissprot and gdb. $gb = new Bio: : DB: : Gen. Bank(); $seq 1 = $gb->get_Seq_by_id('MUSIGHBA 1'); $seq 2 = $gb->get_Seq_by_acc('AF 303112')); $seqio = $gb->get_Stream_by_batch([ qw(J 00522 AF 303112 2981014)]));

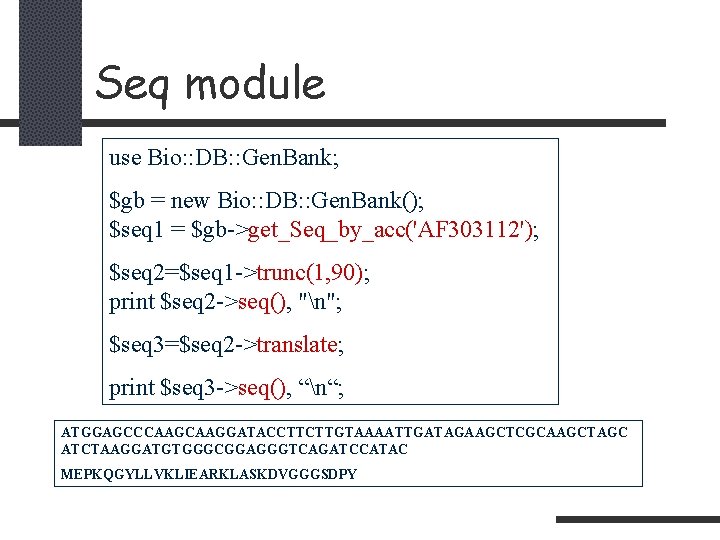
Seq module use Bio: : DB: : Gen. Bank; $gb = new Bio: : DB: : Gen. Bank(); $seq 1 = $gb->get_Seq_by_acc('AF 303112'); $seq 2=$seq 1 ->trunc(1, 90); print $seq 2 ->seq(), "n"; $seq 3=$seq 2 ->translate; print $seq 3 ->seq(), “n“; ATGGAGCCCAAGGATACCTTCTTGTAAAATTGATAGAAGCTCGCAAGCTAGC ATCTAAGGATGTGGGCGGAGGGTCAGATCCATAC MEPKQGYLLVKLIEARKLASKDVGGGSDPY

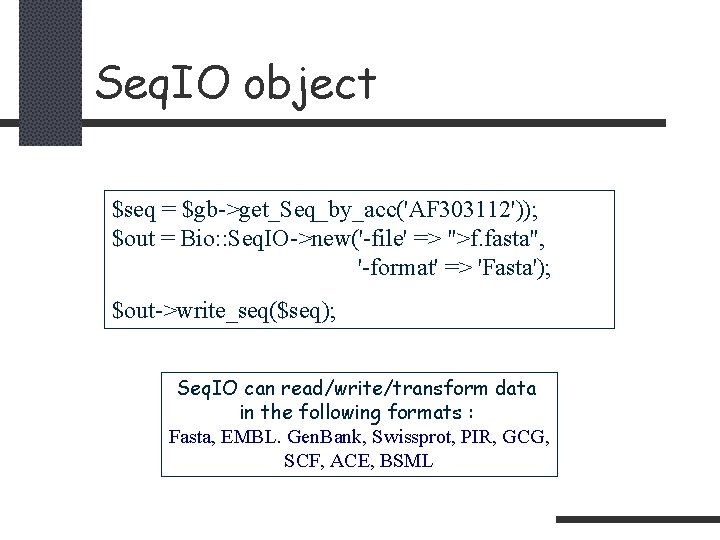
Seq. IO object $seq = $gb->get_Seq_by_acc('AF 303112')); $out = Bio: : Seq. IO->new('-file' => ">f. fasta", '-format' => 'Fasta'); $out->write_seq($seq); Seq. IO can read/write/transform data in the following formats : Fasta, EMBL. Gen. Bank, Swissprot, PIR, GCG, SCF, ACE, BSML

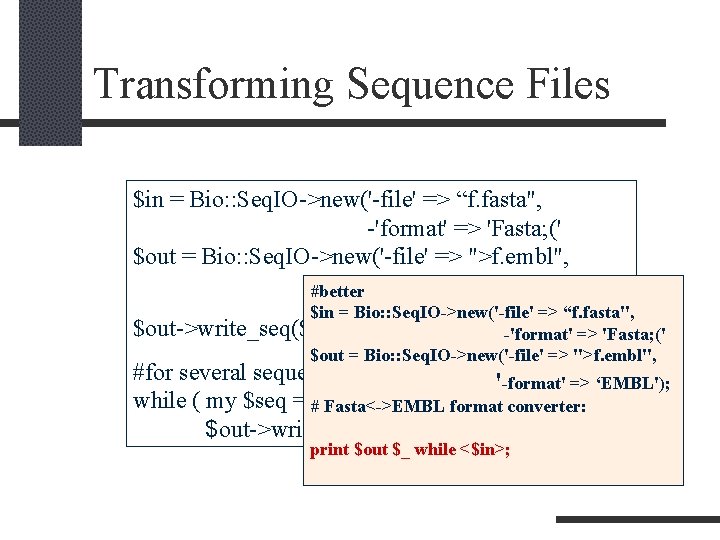
Transforming Sequence Files $in = Bio: : Seq. IO->new('-file' => “f. fasta", -'format' => 'Fasta; (' $out = Bio: : Seq. IO->new('-file' => ">f. embl", #better '-format' => ‘EMBL'); $in = Bio: : Seq. IO->new('-file' => “f. fasta", $out->write_seq($in->next_seq()); -'format' => 'Fasta; (' $out = Bio: : Seq. IO->new('-file' => ">f. embl", #for several sequences '-format' => ‘EMBL'); while ( my $seq = #$in->next_seq() ) { converter: Fasta<->EMBL format $out->write_seq($seq); } print $out $_ while <$in>;

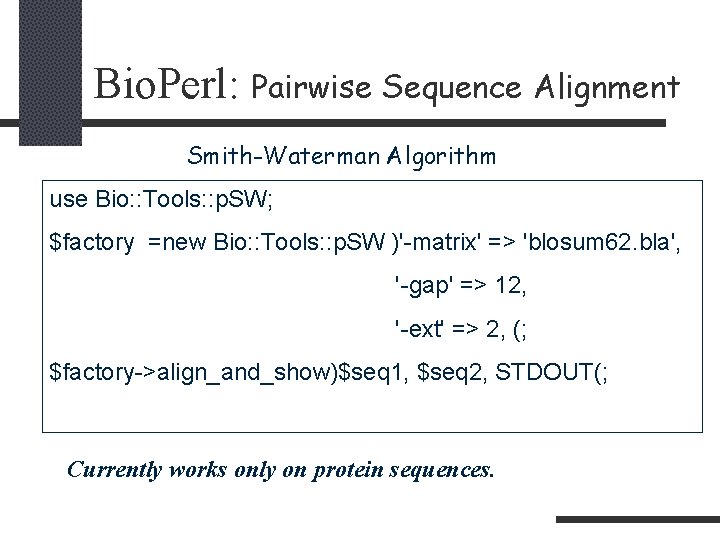
Bio. Perl: Pairwise Sequence Alignment Smith-Waterman Algorithm use Bio: : Tools: : p. SW; $factory =new Bio: : Tools: : p. SW )'-matrix' => 'blosum 62. bla', '-gap' => 12, '-ext' => 2, (; $factory->align_and_show)$seq 1, $seq 2, STDOUT(; Currently works only on protein sequences.

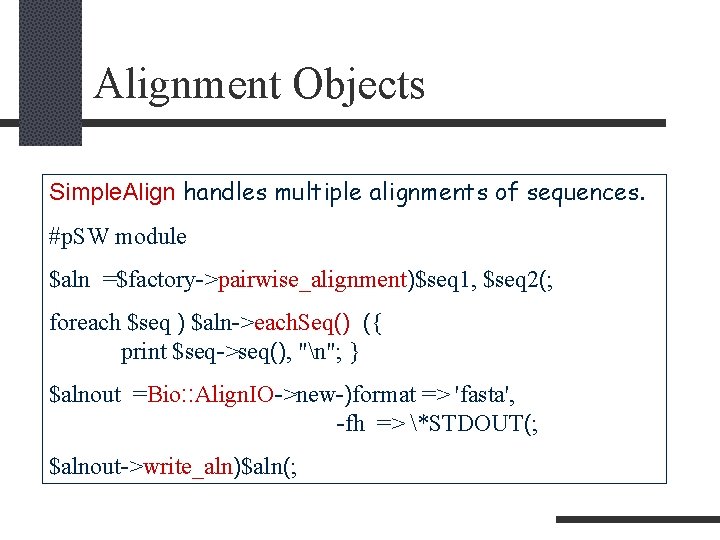
Alignment Objects Simple. Align handles multiple alignments of sequences. #p. SW module $aln =$factory->pairwise_alignment)$seq 1, $seq 2(; foreach $seq ) $aln->each. Seq() ({ print $seq->seq(), "n"; } $alnout =Bio: : Align. IO->new-)format => 'fasta', -fh => *STDOUT(; $alnout->write_aln)$aln(;

Homework Write a cgi script (using Perl) that performs pairwise Local/Global Alignment for DNA sequences. All I/O is via HTML only. Remarks: 1) you are allowed to use only linear space. 1. Choice for Local/Global alignment. 2)To compute z-score perform random shuffling: 2. Two sequences – text boxes or two accession numbers. srand(time| $$); #init, $$-proc. id int(rand($i)); #returns rand. number between [0, $i]. 3. Values for match, mismatch, ins/dels. 3)Shuffling is done in windows (non-overlapping) of 10 4. Number of iterations for computing random scores. bases length. Number of shuffling for each window is Output: random [0, 10]. Input: 1. Alignment score. 2. z-score value (z= (score-average)/standard deviation. )