Sequence Alignment I Dot plots Dynamic Programming Why


![Function dotplot 1. m n function [dotmatrix]=dotplot 1(seq 1, seq 2) ¡ ¡ OUTPUT: Function dotplot 1. m n function [dotmatrix]=dotplot 1(seq 1, seq 2) ¡ ¡ OUTPUT:](https://slidetodoc.com/presentation_image_h2/f8a77fe9123a7636f604200e021b66b0/image-15.jpg)


![Function dotplot 2. m n function [dotmatrix, dot]=dotplot 1(seq 1, seq 2, w, t) Function dotplot 2. m n function [dotmatrix, dot]=dotplot 1(seq 1, seq 2, w, t)](https://slidetodoc.com/presentation_image_h2/f8a77fe9123a7636f604200e021b66b0/image-23.jpg)




















![What is different? Maximization [F(i, j) I(i, j)] = max([F(i-1, j)+g F(i-1, j-1)+w F(i, What is different? Maximization [F(i, j) I(i, j)] = max([F(i-1, j)+g F(i-1, j-1)+w F(i,](https://slidetodoc.com/presentation_image_h2/f8a77fe9123a7636f604200e021b66b0/image-109.jpg)
![What is different? Find the max value of the F matrix [Yj, fj]=max(F)) % What is different? Find the max value of the F matrix [Yj, fj]=max(F)) %](https://slidetodoc.com/presentation_image_h2/f8a77fe9123a7636f604200e021b66b0/image-110.jpg)
- Slides: 111

Sequence Alignment I Dot plots Dynamic Programming

Why align sequences? n conserved sequences conserved function ¡ ¡ ¡ Assess ancestry among homologs (sequences with common ancestory) to help in gene finding and annotation Find consensus motifs among related sequences (e. g. , regulatory and structural regions) Estimate the rate of evolution

Definitions http: //ocw. mit. edu/Ocw. Web/Biology/7 -91 JSpring 2004/Course. Home/

Definitions http: //ocw. mit. edu/Ocw. Web/Biology/7 -91 JSpring 2004/Course. Home/

Problem http: //ocw. mit. edu/Ocw. Web/Biology/7 -91 JSpring 2004/Course. Home/

Are the sequences related? http: //ocw. mit. edu/Ocw. Web/Biology/7 -91 JSpring 2004/Course. Home/

Types of Alignment n n Local Global Gap-free Gapped

Methods of Alignment n n n Dot matrix Dynamic programming K-tuple

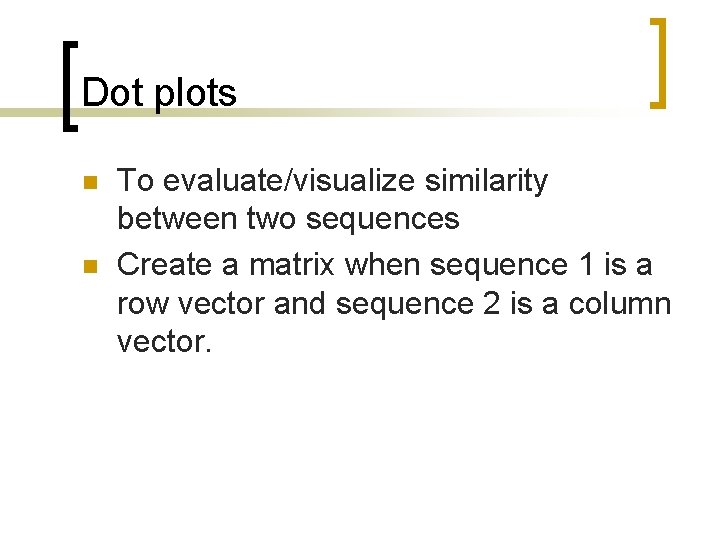
Dot plots n n To evaluate/visualize similarity between two sequences Create a matrix when sequence 1 is a row vector and sequence 2 is a column vector.

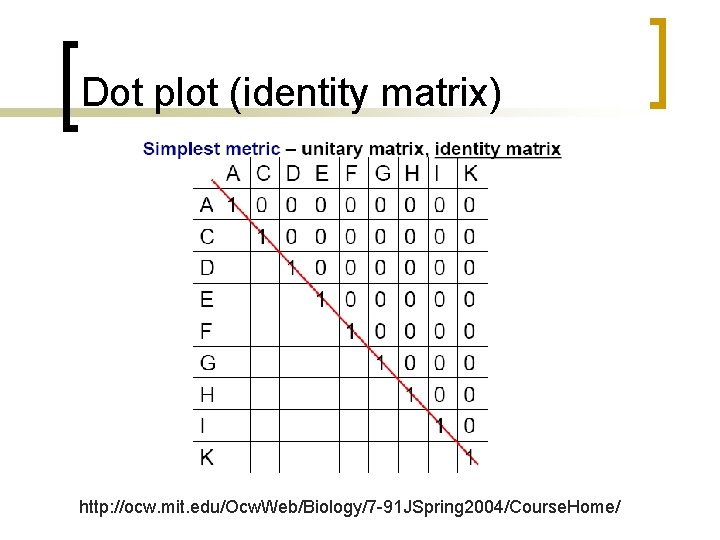
Dot plot (identity matrix) http: //ocw. mit. edu/Ocw. Web/Biology/7 -91 JSpring 2004/Course. Home/

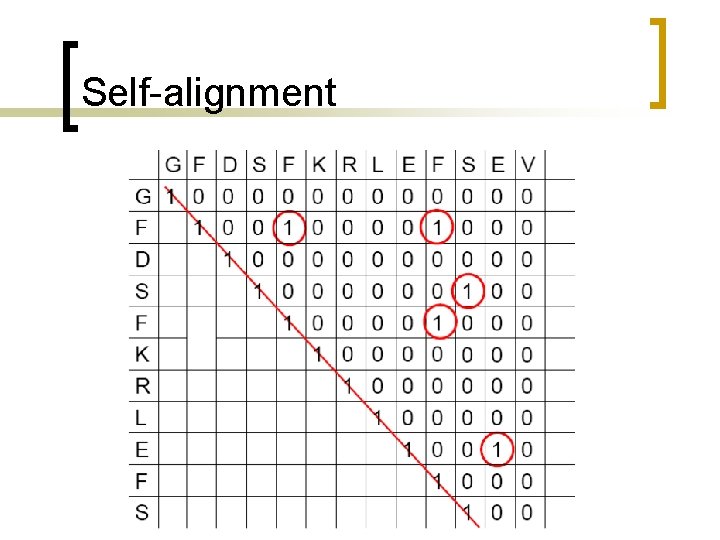
Self-alignment

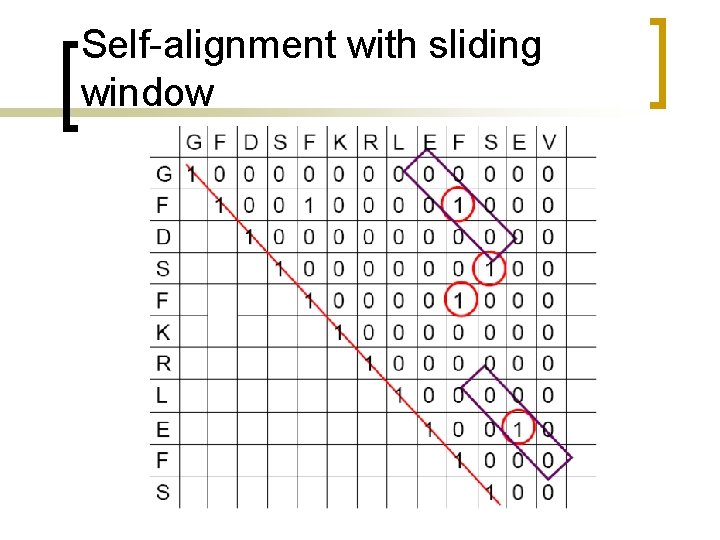
Self-alignment with sliding window

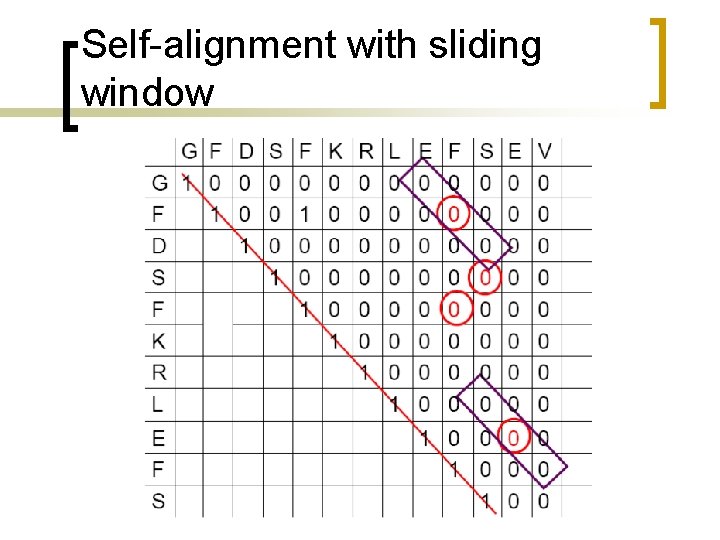
Self-alignment with sliding window

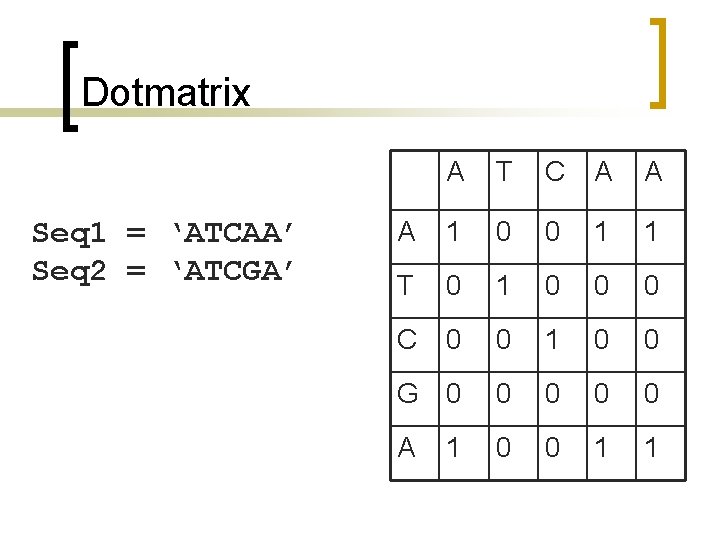
Dotmatrix Seq 1 = ‘ATCAA’ Seq 2 = ‘ATCGA’ A T C A A A 1 0 0 1 1 T 0 1 0 0 0 C 0 0 1 0 0 G 0 0 0 A 0 0 1 1 1
![Function dotplot 1 m n function dotmatrixdotplot 1seq 1 seq 2 OUTPUT Function dotplot 1. m n function [dotmatrix]=dotplot 1(seq 1, seq 2) ¡ ¡ OUTPUT:](https://slidetodoc.com/presentation_image_h2/f8a77fe9123a7636f604200e021b66b0/image-15.jpg)
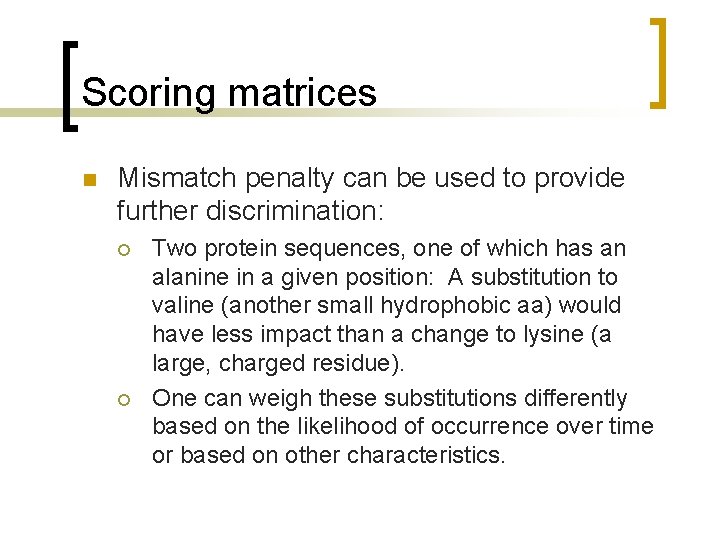
Function dotplot 1. m n function [dotmatrix]=dotplot 1(seq 1, seq 2) ¡ ¡ OUTPUT: [dotmatrix] is an n x m matrix with 1 s and 0 s, where 0 means a mismatch and 1 means a match between two nucleotides INPUT: seq 1 and seq 2 are strings made of a, t, c, or g. (seq 1 is a row vector 1 x m; and seq 2 is a column vector (n x 1).

Run dotplot 1. m n n Place dotplot 1. m into your directory. Open a Matlab session. Open the file named dotplot 1. m under the file option. Define seq 1 and seq 2 as the following; and run the program: >> seq 1 = ‘attataagg’ >> seq 2 = ‘attataggg’ >> [dotmatrix] = dotplot 1(seq 1, seq 2) n Next, copy and paste each line of the dotplot 1. m on the command window without using ; to track what the program is doing.

Dot plot Seq 1='attataagg‘ Seq 2='attataggg' A match is red (=1); a mismatch is blue (=0)

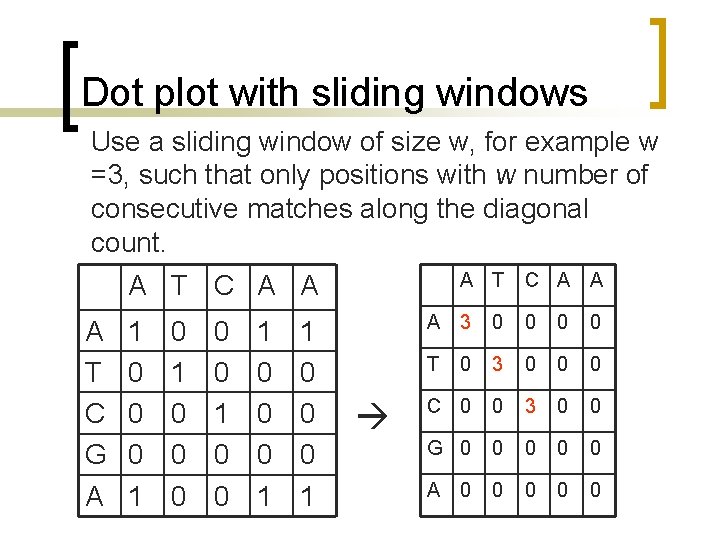
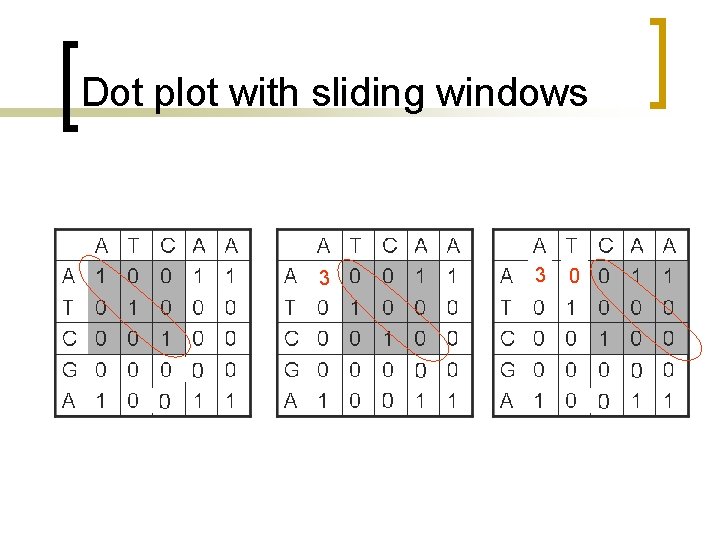
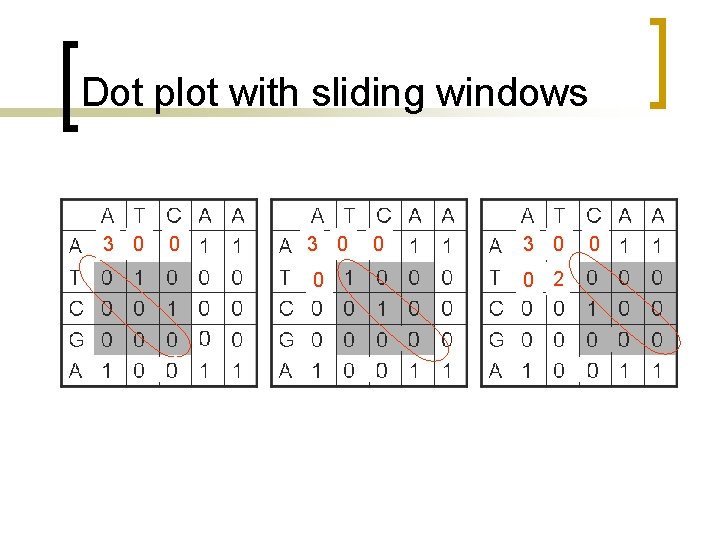
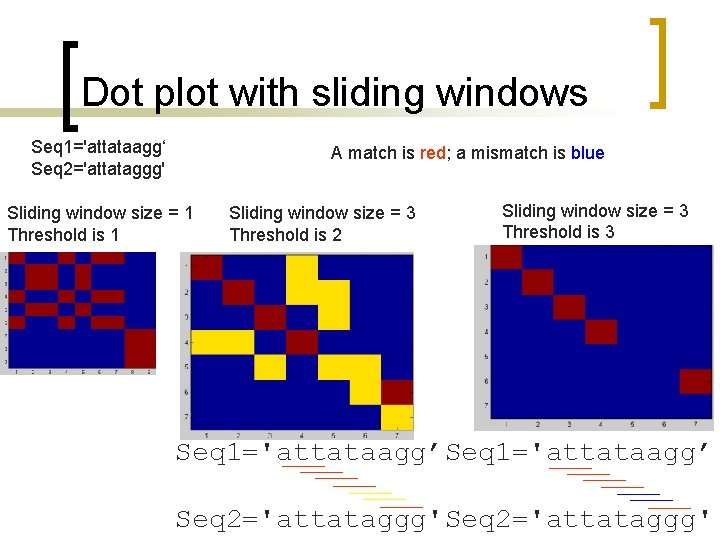
Dot plot with sliding windows n n n Sliding windows consider more than one position at a time. Similarity cutoffs (threshold) also allow to get rid of positions with low similarity across the window. Sliding window can reduce the noise in the dot plot.

Dot plot with sliding windows Use a sliding window of size w, for example w =3, such that only positions with w number of consecutive matches along the diagonal count. A T C A A A T C G A 1 0 0 0 1 0 0 0 1 C A A A 3 0 0 T 0 3 0 0 0 C 0 0 3 0 0 G 0 0 0 A 0 0 0

Dot plot with sliding windows 3 3 0

Dot plot with sliding windows 3 0 0 0 3 0 0 2 0

Dot plot with sliding windows A T C A A A 3 0 0 T 0 2 0 0 0 C 0 0 2 0 0 G 0 0 0 A 0 0 0
![Function dotplot 2 m n function dotmatrix dotdotplot 1seq 1 seq 2 w t Function dotplot 2. m n function [dotmatrix, dot]=dotplot 1(seq 1, seq 2, w, t)](https://slidetodoc.com/presentation_image_h2/f8a77fe9123a7636f604200e021b66b0/image-23.jpg)
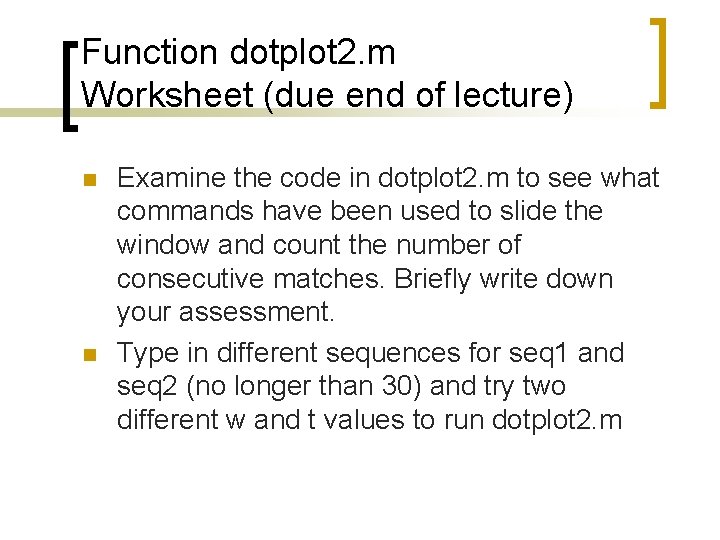
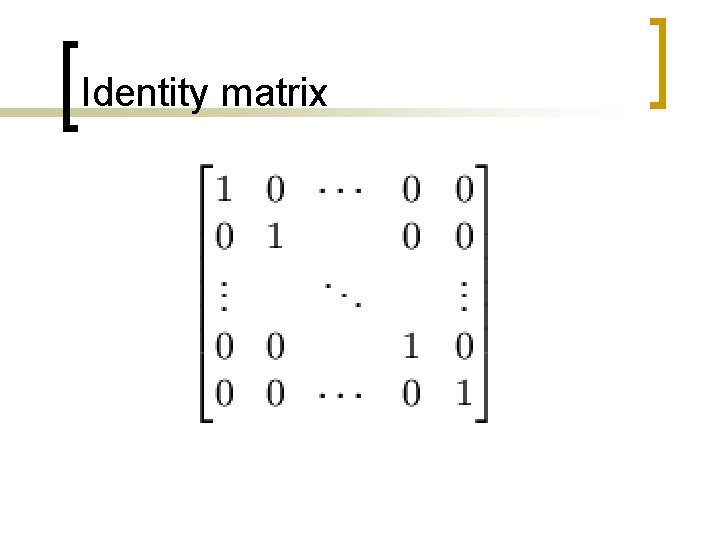
Function dotplot 2. m n function [dotmatrix, dot]=dotplot 1(seq 1, seq 2, w, t) n Introduced two variables: ¡ ¡ W = size of the sliding window T = threshold, number of matches along the diagonal to assume a match.

Function dotplot 2. m Worksheet (due end of lecture) n n Examine the code in dotplot 2. m to see what commands have been used to slide the window and count the number of consecutive matches. Briefly write down your assessment. Type in different sequences for seq 1 and seq 2 (no longer than 30) and try two different w and t values to run dotplot 2. m

Dot plot with sliding windows Seq 1='attataagg‘ Seq 2='attataggg' A match is red; a mismatch is blue Sliding window size = 1 Threshold is 1 Sliding window size = 3 Threshold is 2 Sliding window size = 3 Threshold is 3 Seq 1='attataagg’ Seq 2='attataggg'

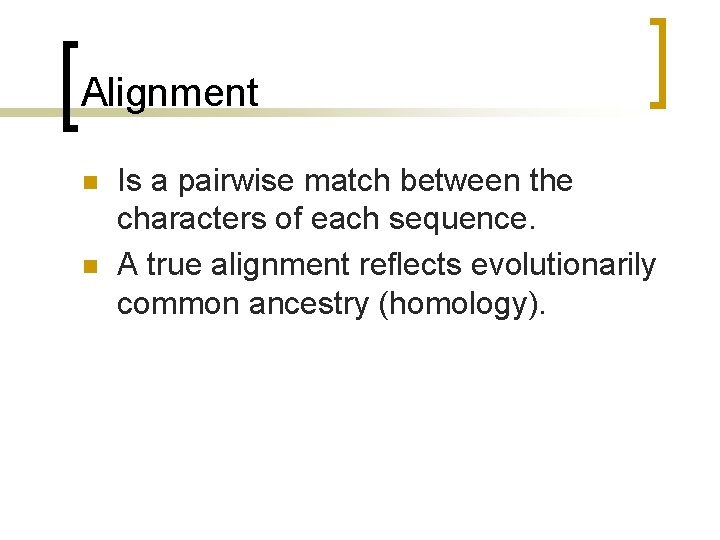
Alignment n n Is a pairwise match between the characters of each sequence. A true alignment reflects evolutionarily common ancestry (homology).

Changes that occur in sequences n n A mutation that replaces one character with another is a substitution. An insertion that adds one or more positions and a deletion that deletes one or more positions are known as indels (gaps)

Alignment example: n n n AATCTATA AAGATA

Gap-free alignment: match and mismatch n n An alignment receives for each aligned pair of identical residues (the match score) and the penalty for aligned pair of nonidentical residues (mismatch score). match score; if seq 1=seq 2 mismatch score; if seq 1 seq 2 nwhere n is the length of the longer sequence.

Gap-free alignment example: n n n AATCTATA AAGATA Alignment scores would be 4, 1, 3, respectively; if the match score is 1 and mismatch score is 0.

Gaps n Indels complicate alignments by increasing the number of possible alignments between two or more sequences.

Alignment: match, mismatch, and gap penalty n n An alignment receives a score for each aligned pair of identical residues (the match score) and the penalty for aligned pair of nonidentical residues (mismatch score), and a penalty for insertion of gaps. gap penalty; if seq 1 = ‘-’ or seq 2 = ‘-’ match score; if no gaps and seq 1=seq 2 mismatch score; if no gaps and seq 1 seq 2 nwhere n is the length of the longer sequence.

Gap-free alignment example: n n n AATCTATA AAG-AT-A AATCTATA AA-G-ATA AATCTATA AA--GATA Alignment scores would be 1, 3, 3, respectively; if the match score is 1; mismatch score is 0; and gap penalty is -1.

Origination and Length Penalties n n Simple gap penalties lead to many optimal alignments (those having the same score). Mutations are rare, invoking fewest number of unlikely events is evolutionarily sound. ¡ 3 -nt indel would be more common than multiple single indels.

Origination and Length Penalties n n Origination penalty: starting a new series of gaps in one of the sequences being aligned. Length penalty: number of sequential missing characters.

Gap-free alignment example: n n n AATCTATA AAG-AT-A AATCTATA AA-G-ATA AATCTATA AA--GATA Alignment scores would be -3, -1, +1, respectively; if the match score is 1; mismatch score is 0; and origination penalty is -2 and length penalty is -1.

Scoring matrices n Mismatch penalty can be used to provide further discrimination: ¡ ¡ Two protein sequences, one of which has an alanine in a given position: A substitution to valine (another small hydrophobic aa) would have less impact than a change to lysine (a large, charged residue). One can weigh these substitutions differently based on the likelihood of occurrence over time or based on other characteristics.

Scoring matrices n A scoring matrix is used to score each nongap position in the alignment. ¡ ¡ Nucleotide sequences Amino acid sequences

Identity matrix

Scoring matrices for DNA sequences A T C G A 5 -4 -4 -4 A 1 -5 -5 -1 T -4 5 -4 -4 T -5 1 -1 -5 C -4 -4 5 -4 C -5 -1 1 -5 G -4 5 G -1 -5 -5 1 BLAST matrix Transition/Transversion matrix

Scoring Matrices Need to know the frequency of one amino acid substituting for another versus that event happening by chance alone based on frequency of occurrence of each amino acid: odds ratio = P(ab)/q(a)*q(b)

Scoring matrices for amino acids n n Blosum PAM (point accepted mutation)

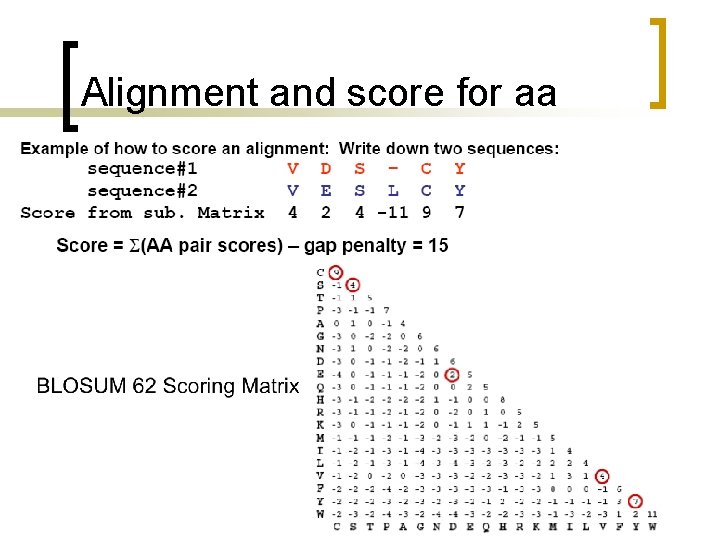
Alignment and score for aa

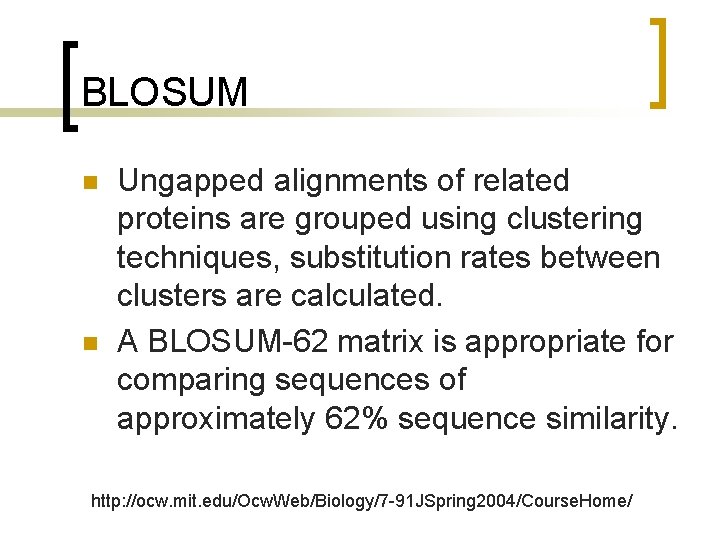
BLOSUM n n Ungapped alignments of related proteins are grouped using clustering techniques, substitution rates between clusters are calculated. A BLOSUM-62 matrix is appropriate for comparing sequences of approximately 62% sequence similarity. http: //ocw. mit. edu/Ocw. Web/Biology/7 -91 JSpring 2004/Course. Home/

PAM matrices http: //ocw. mit. edu/Ocw. Web/Biology/7 -91 JSpring 2004/Course. Home/

PAM matrices http: //ocw. mit. edu/Ocw. Web/Biology/7 -91 JSpring 2004/Course. Home/

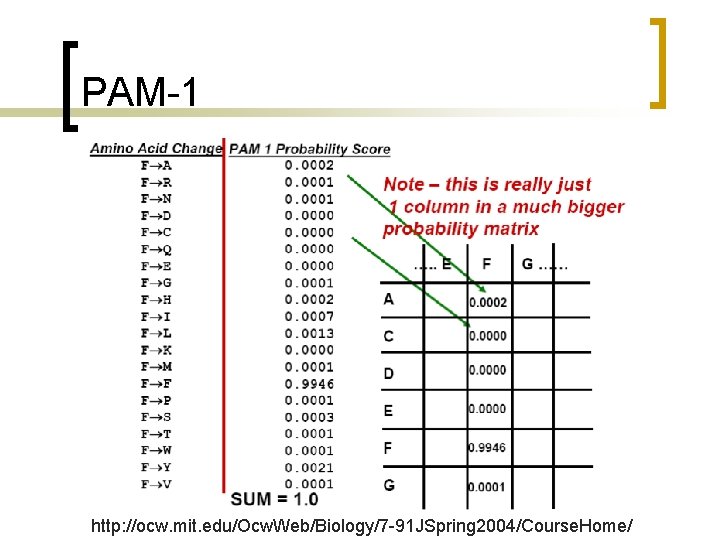
PAM-1 http: //ocw. mit. edu/Ocw. Web/Biology/7 -91 JSpring 2004/Course. Home/

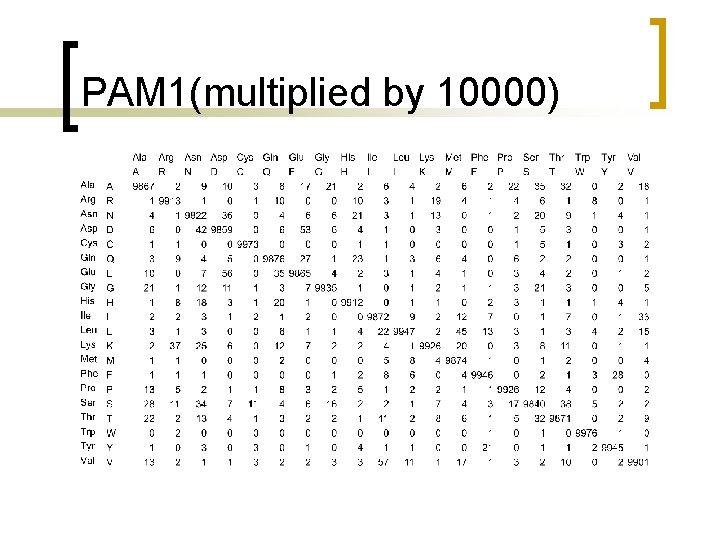
PAM 1(multiplied by 10000)

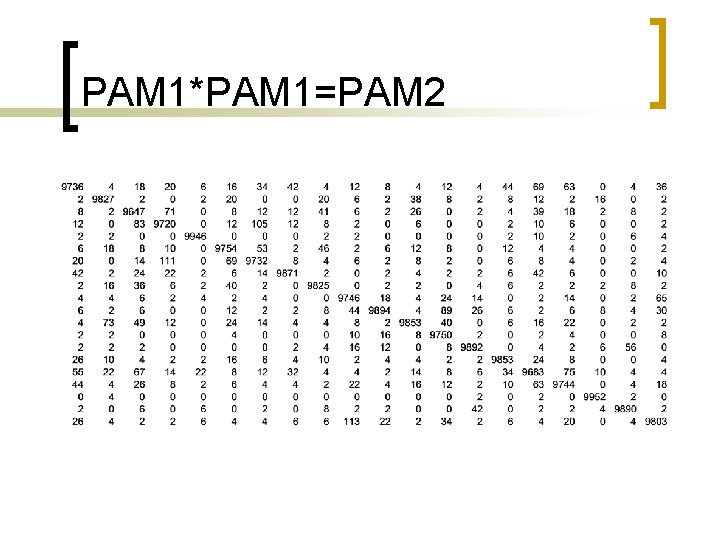
PAM 1*PAM 1=PAM 2

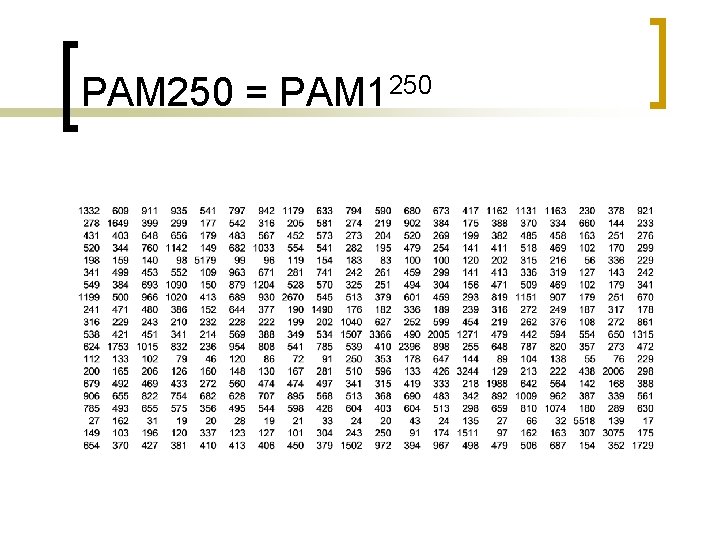
PAM 250 = PAM 1250

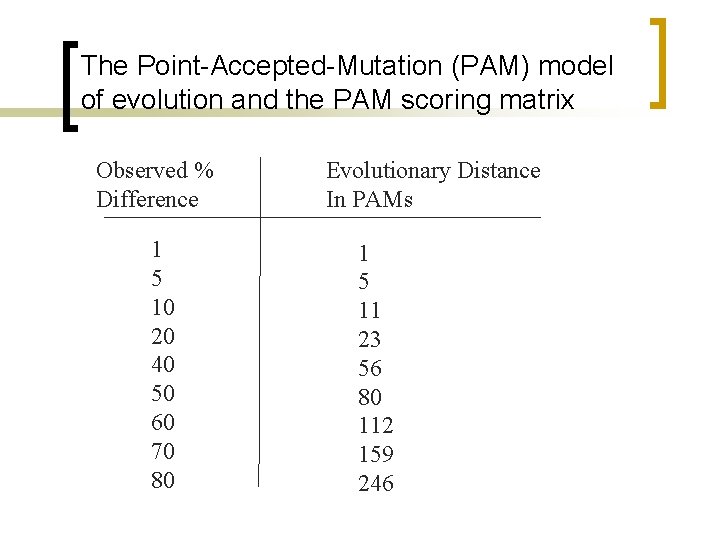
The Point-Accepted-Mutation (PAM) model of evolution and the PAM scoring matrix Observed % Difference 1 5 10 20 40 50 60 70 80 Evolutionary Distance In PAMs 1 5 11 23 56 80 112 159 246

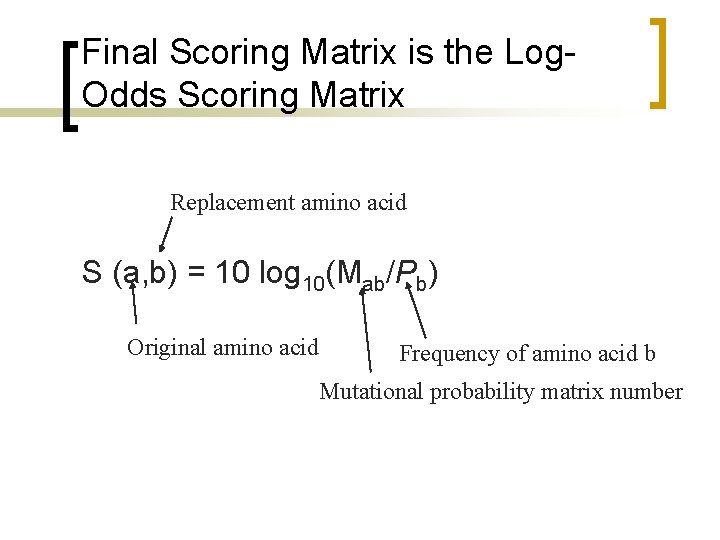
Final Scoring Matrix is the Log. Odds Scoring Matrix Replacement amino acid S (a, b) = 10 log 10(Mab/Pb) Original amino acid Frequency of amino acid b Mutational probability matrix number

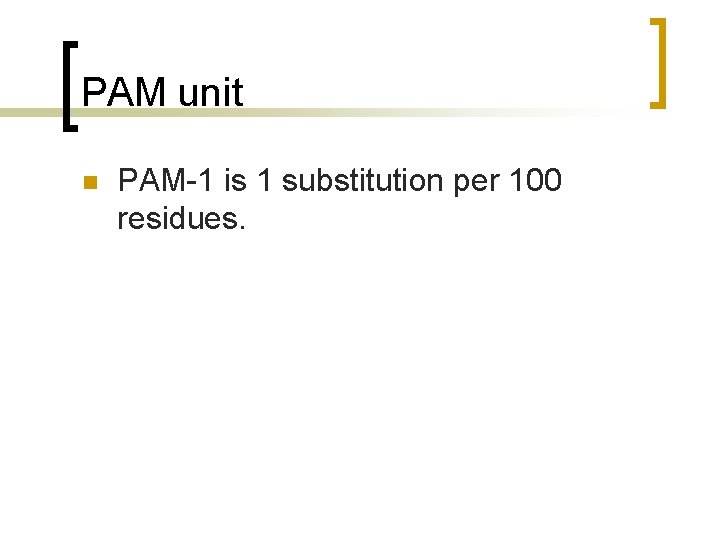
PAM unit n PAM-1 is 1 substitution per 100 residues.

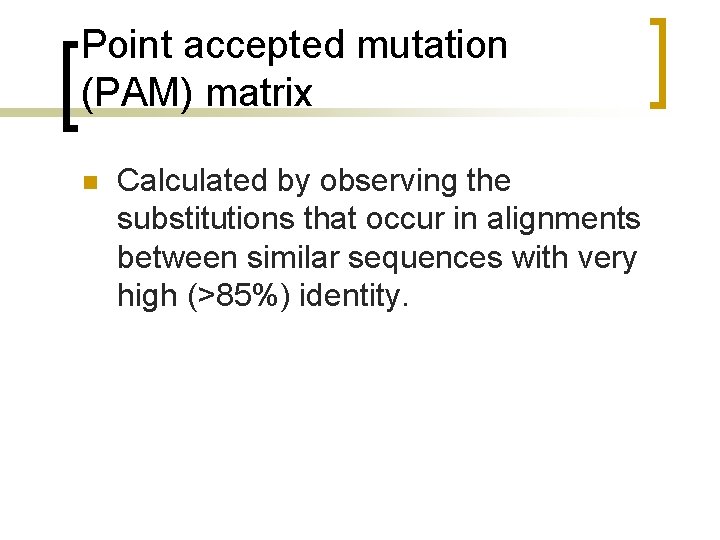
Point accepted mutation (PAM) matrix n Calculated by observing the substitutions that occur in alignments between similar sequences with very high (>85%) identity.

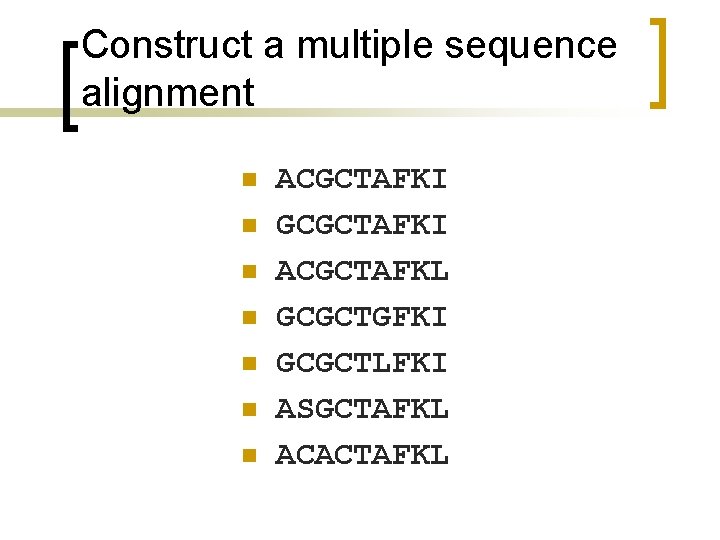
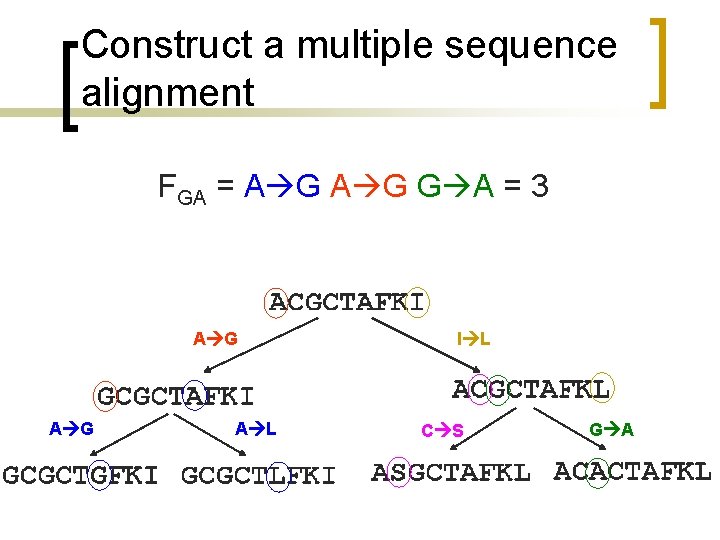
Construct a multiple sequence alignment n n n n ACGCTAFKI GCGCTAFKI ACGCTAFKL GCGCTGFKI GCGCTLFKI ASGCTAFKL ACACTAFKL

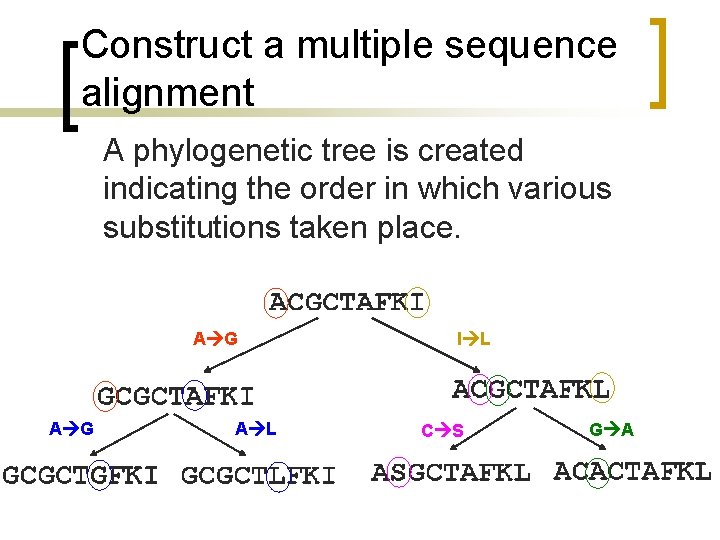
Construct a multiple sequence alignment A phylogenetic tree is created indicating the order in which various substitutions taken place. A G A L I L C S G A

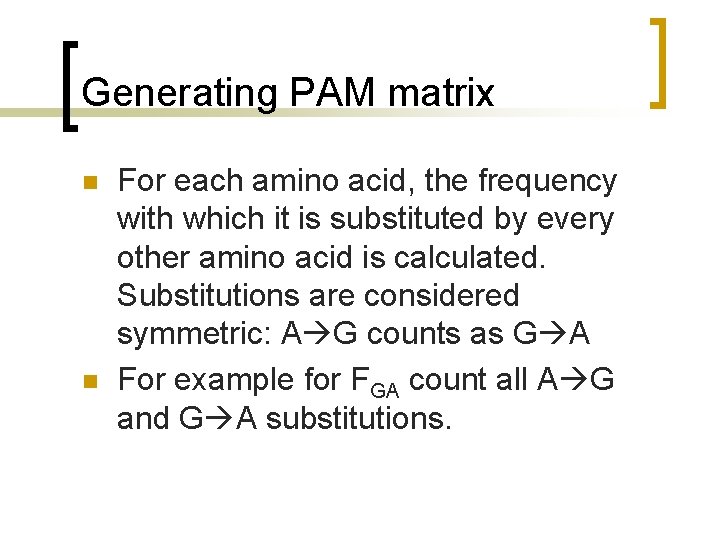
Generating PAM matrix n n For each amino acid, the frequency with which it is substituted by every other amino acid is calculated. Substitutions are considered symmetric: A G counts as G A For example for FGA count all A G and G A substitutions.

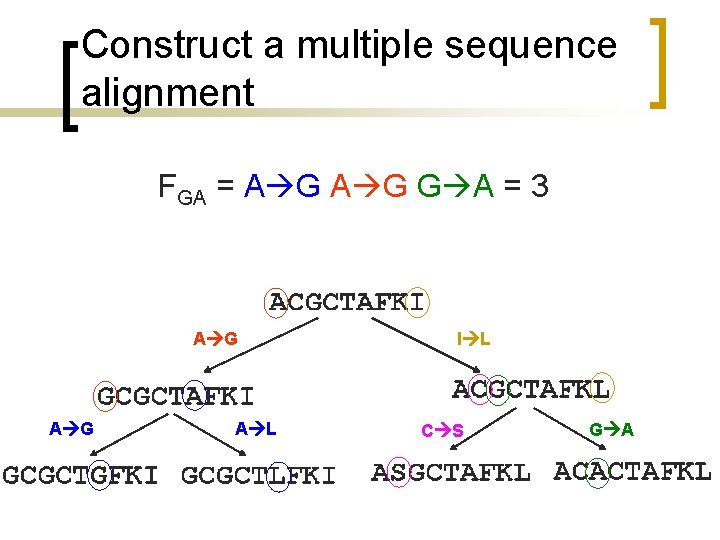
Construct a multiple sequence alignment FGA = A G G A = 3 A G A L I L C S G A

Relative mutability, mi n n n Number of times the amino acid is substituted by any other amino acid in the phylogenetic tree. This number is then divided by the total number of mutations that could have affected the residue. This denominator is the total number of subs across entire tree times two, multiplied by freq. of the amino acid, times a scaling factor

Construct a multiple sequence alignment FGA = A G G A = 3 A G A L I L C S G A

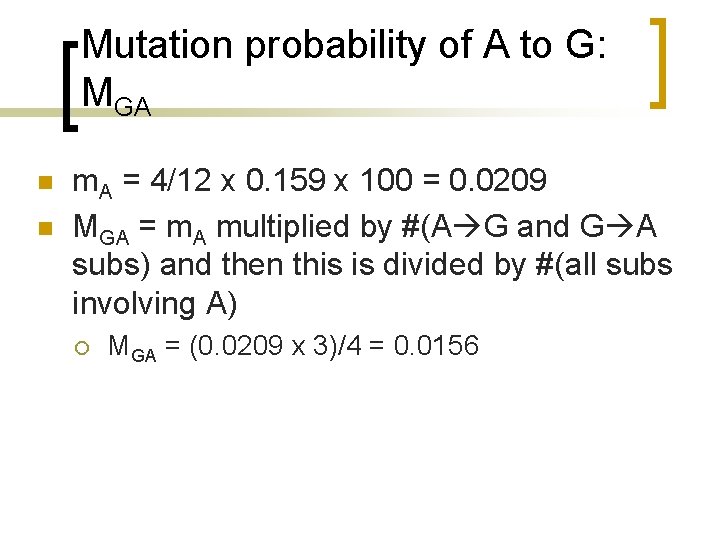
Relative mutability of A: m. A n n Total of 4 mutations involving A. Total number mutations in the entire tree is 6 should be multiplied by two = 6 x 2 = 12 Relative frequency of A residues (10 As out of 7 x 9=63 residues) is 10/63 = 0. 159. Thus m. A = 4/12 x 0. 159 x 100 = 0. 0209

Mutation probability of A to G: MGA n n m. A = 4/12 x 0. 159 x 100 = 0. 0209 MGA = m. A multiplied by #(A G and G A subs) and then this is divided by #(all subs involving A) ¡ MGA = (0. 0209 x 3)/4 = 0. 0156

Scoring matrix: RGA n n log(Mij/fi), where Mij mutation probability of ij and fi equals to relative frequency of j type residue. For example f(G) = 10/63 = 0. 1587 RGA = log(MGA/f(G)) = log(0. 0156/0. 1587) = -1. 01

PAM matrix calculator n http: //www. bioinformatics. nl/tools/pam. html

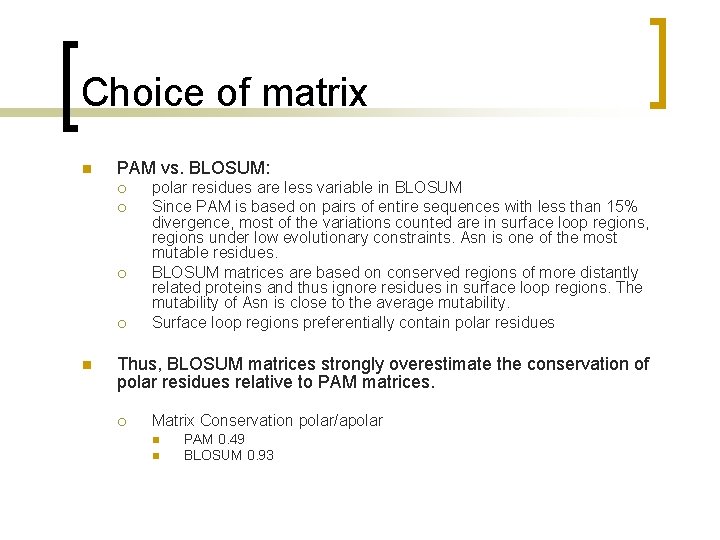
Choice of matrix n PAM vs. BLOSUM: ¡ ¡ n polar residues are less variable in BLOSUM Since PAM is based on pairs of entire sequences with less than 15% divergence, most of the variations counted are in surface loop regions, regions under low evolutionary constraints. Asn is one of the most mutable residues. BLOSUM matrices are based on conserved regions of more distantly related proteins and thus ignore residues in surface loop regions. The mutability of Asn is close to the average mutability. Surface loop regions preferentially contain polar residues Thus, BLOSUM matrices strongly overestimate the conservation of polar residues relative to PAM matrices. ¡ Matrix Conservation polar/apolar n n PAM 0. 49 BLOSUM 0. 93

Choice of matrix n General: the choice of matrix should be governed by the use to which the matrix is put. ¡ Searches for distantly similar sequences, in which only the BLOCKS are likely to be conserved should rely on BLOSUM matrices. ¡ Complete alignments of globular protein sequences should use PAM matrices. ¡ Proteins with extensive transmembrane helices should be aligned using the transmembrane matrix.

Choice of matrix http: //mcb. berkeley. edu/labs/king/blast/docs/matrix_info. html

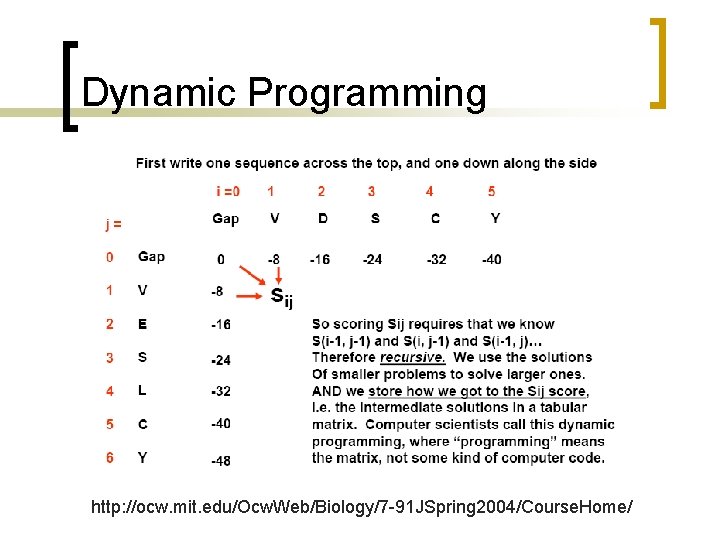
Dynamic Programming n n Exhaustive search: search all possible alignments: NOT FEASIBLE Dynamic programming: a method of breaking a problem apart into reasonably sized subproblems and using these partial results to compute the final answer. ¡ Needleman and Wunsch

How to start the alignment? n How to break down the problem: ¡ ¡ Seq 1 = ‘CACGA’ Seq 2 = ‘CGA’ n Three possible ways to start the problem: ¡ ¡ ¡ C of seq 1 and C of seq 2 match C ACGA C GA A gap is inserted into 1 st position of seq 1 CACGA C GA A gap is inserted into 1 st position of seq 2 C ACGA

How to end the alignment? n If we knew the score for the best alignment, we can track back the steps to reach to the best score. ¡ ¡ Use a table to store each step of the alignment to refer back later on. Depends on storing partial sequence alignments so you don’t do it again and again.

Dynamic Programming

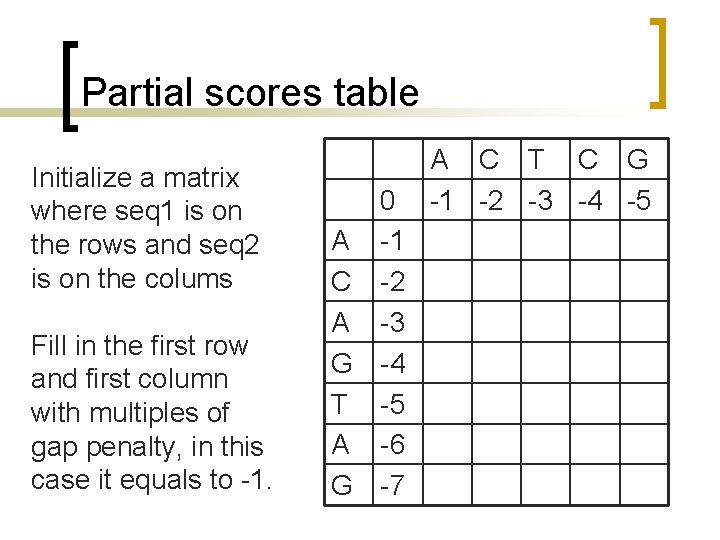
Partial scores table Initialize a matrix where seq 1 is on the rows and seq 2 is on the colums Fill in the first row and first column with multiples of gap penalty, in this case it equals to -1. 0 A -1 C -2 A -3 G -4 T -5 A -6 G -7 A C T C G -1 -2 -3 -4 -5

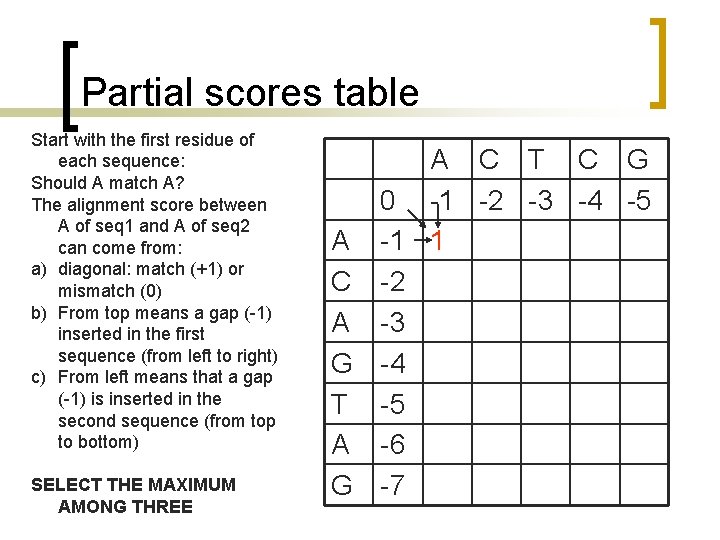
Partial scores table Start with the first residue of each sequence: Should A match A? The alignment score between A of seq 1 and A of seq 2 can come from: a) diagonal: match (+1) or mismatch (0) b) From top means a gap (-1) inserted in the first sequence (from left to right) c) From left means that a gap (-1) is inserted in the second sequence (from top to bottom) SELECT THE MAXIMUM AMONG THREE A C T C G 0 -1 -2 -3 -4 -5 A -1 1 C -2 A -3 G -4 T -5 A -6 G -7

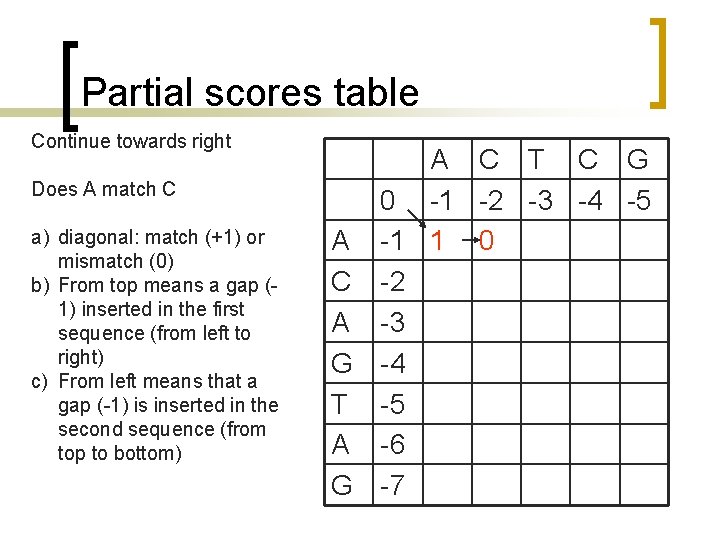
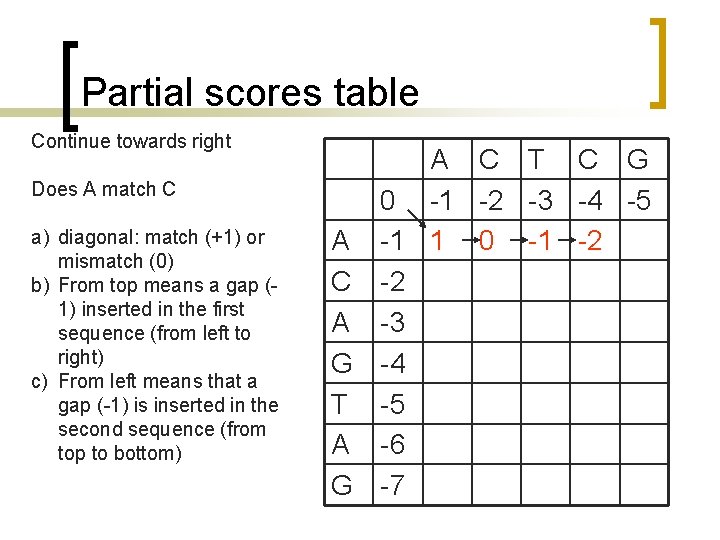
Partial scores table Continue towards right Does A match C a) diagonal: match (+1) or mismatch (0) b) From top means a gap (1) inserted in the first sequence (from left to right) c) From left means that a gap (-1) is inserted in the second sequence (from top to bottom) A C T C G 0 -1 -2 -3 -4 -5 A -1 1 0 C -2 A -3 G -4 T -5 A -6 G -7

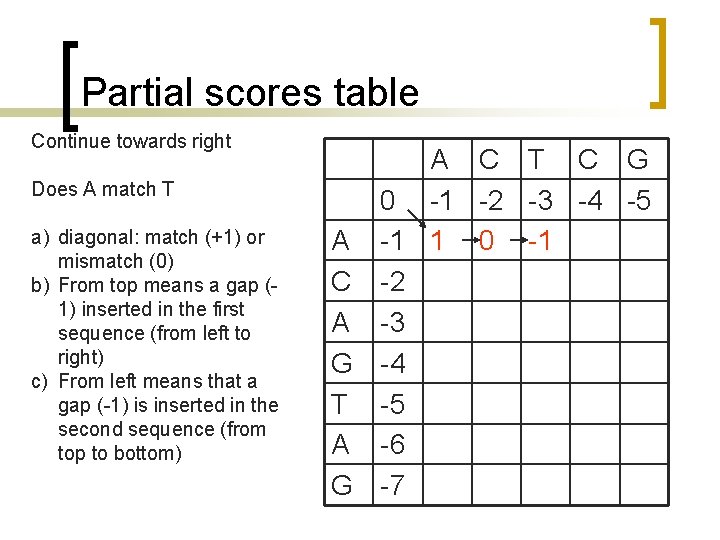
Partial scores table Continue towards right Does A match T a) diagonal: match (+1) or mismatch (0) b) From top means a gap (1) inserted in the first sequence (from left to right) c) From left means that a gap (-1) is inserted in the second sequence (from top to bottom) A C T C G 0 -1 -2 -3 -4 -5 A -1 1 0 -1 C -2 A -3 G -4 T -5 A -6 G -7

Partial scores table Continue towards right Does A match C a) diagonal: match (+1) or mismatch (0) b) From top means a gap (1) inserted in the first sequence (from left to right) c) From left means that a gap (-1) is inserted in the second sequence (from top to bottom) A C T C G 0 -1 -2 -3 -4 -5 A -1 1 0 -1 -2 C -2 A -3 G -4 T -5 A -6 G -7

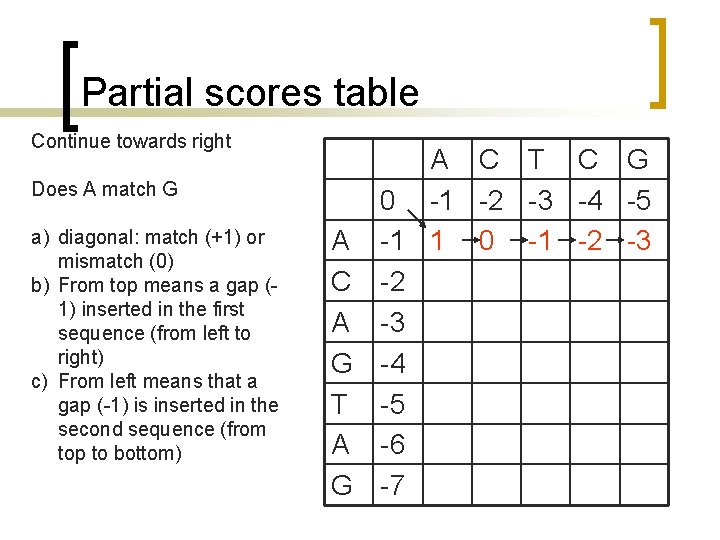
Partial scores table Continue towards right Does A match G a) diagonal: match (+1) or mismatch (0) b) From top means a gap (1) inserted in the first sequence (from left to right) c) From left means that a gap (-1) is inserted in the second sequence (from top to bottom) A C T C G 0 -1 -2 -3 -4 -5 A -1 1 0 -1 -2 -3 C -2 A -3 G -4 T -5 A -6 G -7

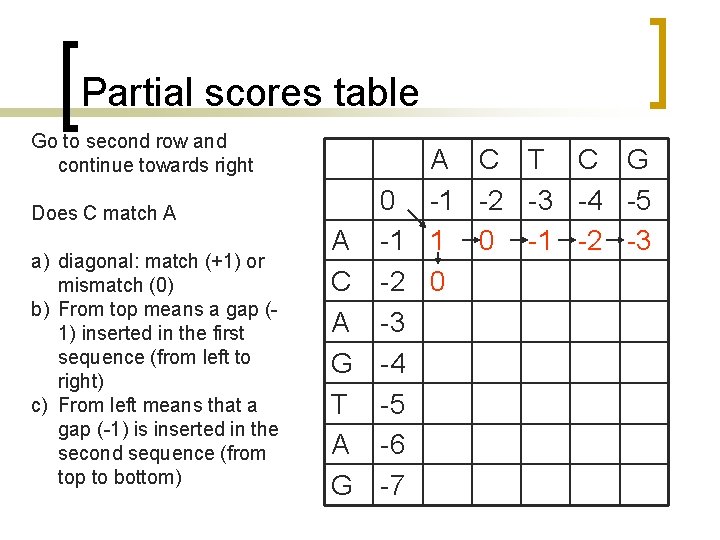
Partial scores table Go to second row and continue towards right Does C match A a) diagonal: match (+1) or mismatch (0) b) From top means a gap (1) inserted in the first sequence (from left to right) c) From left means that a gap (-1) is inserted in the second sequence (from top to bottom) A C T C G 0 -1 -2 -3 -4 -5 A -1 1 0 -1 -2 -3 C -2 0 A -3 G -4 T -5 A -6 G -7

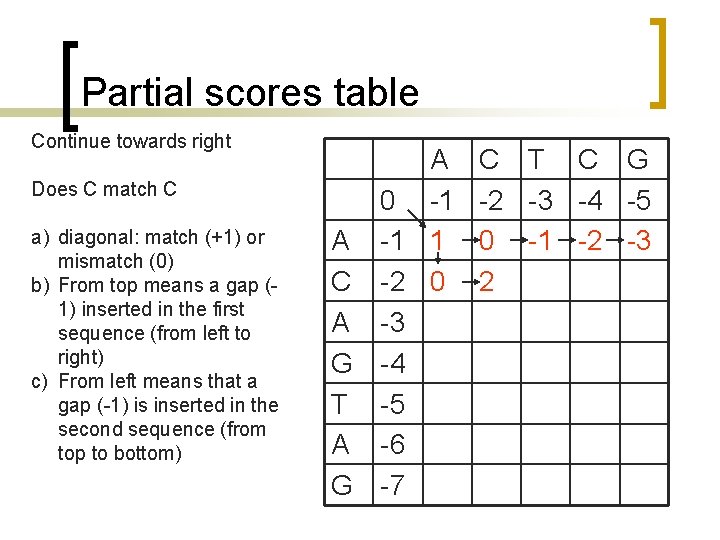
Partial scores table Continue towards right Does C match C a) diagonal: match (+1) or mismatch (0) b) From top means a gap (1) inserted in the first sequence (from left to right) c) From left means that a gap (-1) is inserted in the second sequence (from top to bottom) A 0 -1 A -1 1 C -2 0 A -3 G -4 T -5 A -6 G -7 C T C G -2 -3 -4 -5 0 -1 -2 -3 2

Partial scores table Continue towards right Does C match T a) diagonal: match (+1) or mismatch (0) b) From top means a gap (1) inserted in the first sequence (from left to right) c) From left means that a gap (-1) is inserted in the second sequence (from top to bottom) A 0 -1 A -1 1 C -2 0 A -3 G -4 T -5 A -6 G -7 C -2 0 2 T C G -3 -4 -5 -1 -2 -3 1

Partial scores table Continue towards right Does C match C a) diagonal: match (+1) or mismatch (0) b) From top means a gap (1) inserted in the first sequence (from left to right) c) From left means that a gap (-1) is inserted in the second sequence (from top to bottom) A 0 -1 A -1 1 C -2 0 A -3 G -4 T -5 A -6 G -7 C -2 0 2 T -3 -1 1 C G -4 -5 -2 -3 0

Partial scores table Continue towards right Does C match G a) diagonal: match (+1) or mismatch (0) b) From top means a gap (1) inserted in the first sequence (from left to right) c) From left means that a gap (-1) is inserted in the second sequence (from top to bottom) A 0 -1 A -1 1 C -2 0 A -3 G -4 T -5 A -6 G -7 C -2 0 2 T -3 -1 1 C -4 -2 0 G -5 -3 -1

Partial scores table Fill in all cells in the partial scores table While you fill in keep tract of from which direction you have carried away the scores from. 0 A -1 C -2 A -3 G -4 T -5 A -6 G -7 A -1 1 0 -1 -2 -3 -4 -5 C -2 0 2 1 0 -1 -2 -3 T -3 -1 1 2 1 1 0 -1 C -4 -2 0 1 2 1 1 0 G -5 -3 -1 0 2 2 1 2

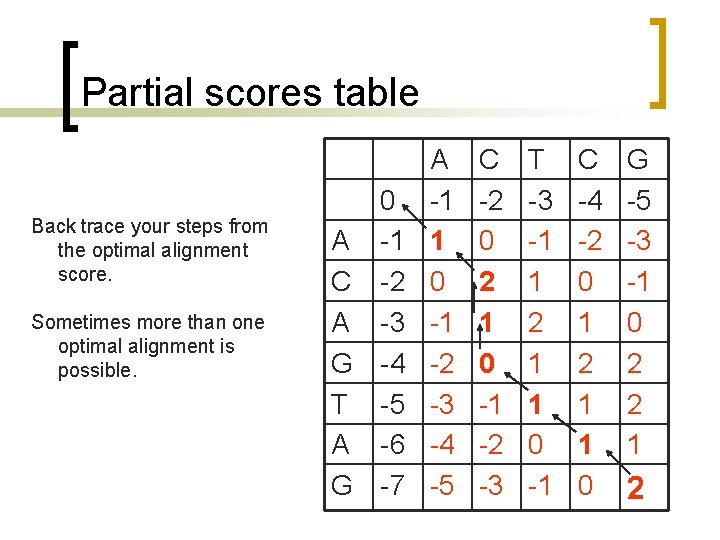
Partial scores table Back trace your steps from the optimal alignment score. Sometimes more than one optimal alignment is possible. 0 A -1 C -2 A -3 G -4 T -5 A -6 G -7 A -1 1 0 -1 -2 -3 -4 -5 C -2 0 2 1 0 -1 -2 -3 T -3 -1 1 2 1 1 0 -1 C -4 -2 0 1 2 1 1 0 G -5 -3 -1 0 2 2 1 2

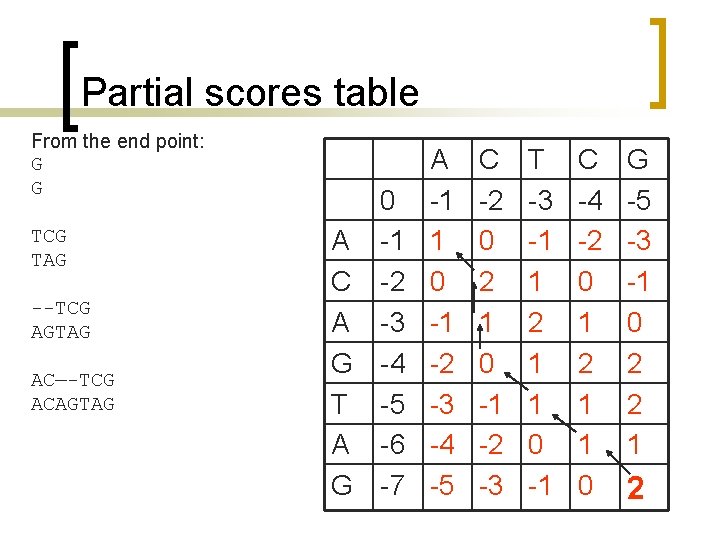
Partial scores table From the end point: G G TCG TAG --TCG AGTAG AC—-TCG ACAGTAG 0 A -1 C -2 A -3 G -4 T -5 A -6 G -7 A -1 1 0 -1 -2 -3 -4 -5 C -2 0 2 1 0 -1 -2 -3 T -3 -1 1 2 1 1 0 -1 C -4 -2 0 1 2 1 1 0 G -5 -3 -1 0 2 2 1 2

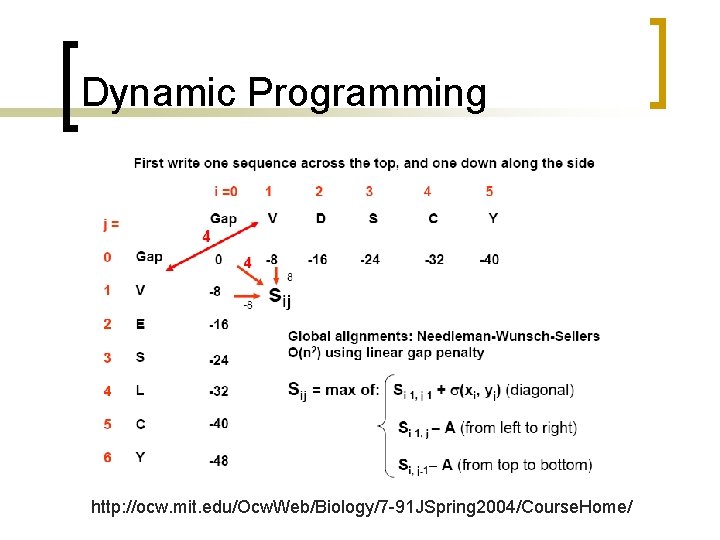
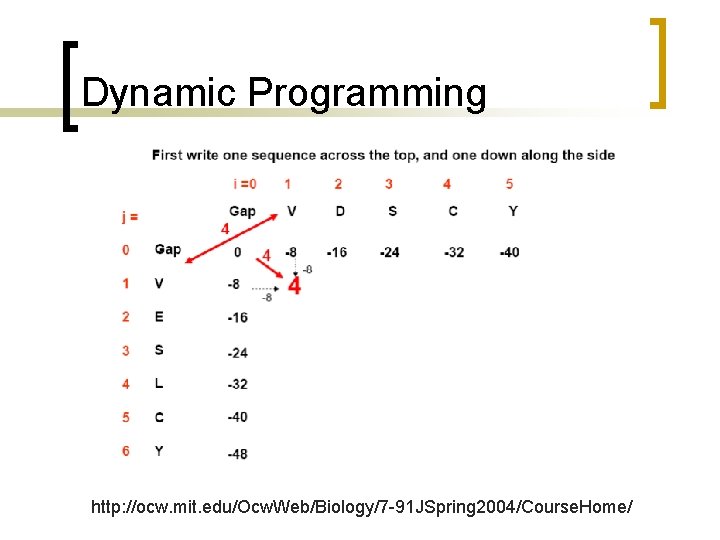
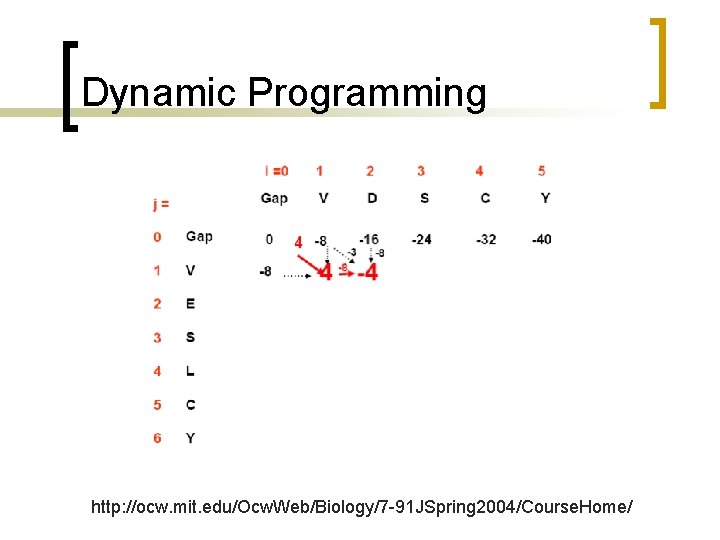
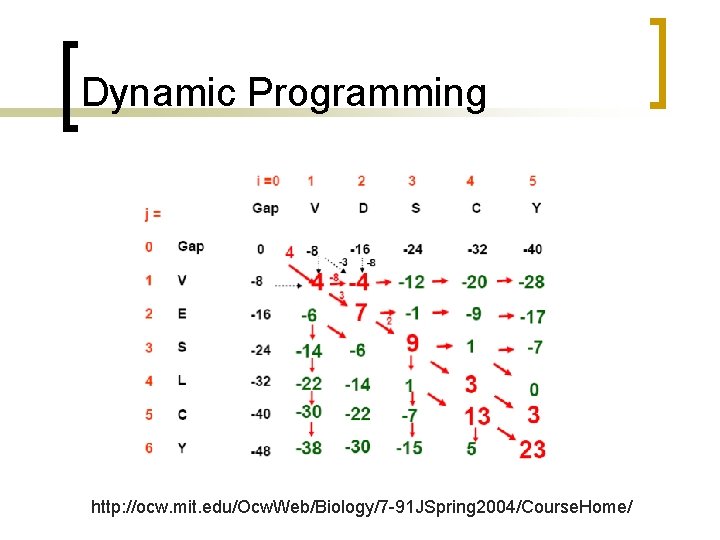
Dynamic Programming http: //ocw. mit. edu/Ocw. Web/Biology/7 -91 JSpring 2004/Course. Home/

Dynamic Programming http: //ocw. mit. edu/Ocw. Web/Biology/7 -91 JSpring 2004/Course. Home/

Dynamic Programming http: //ocw. mit. edu/Ocw. Web/Biology/7 -91 JSpring 2004/Course. Home/

Dynamic Programming http: //ocw. mit. edu/Ocw. Web/Biology/7 -91 JSpring 2004/Course. Home/

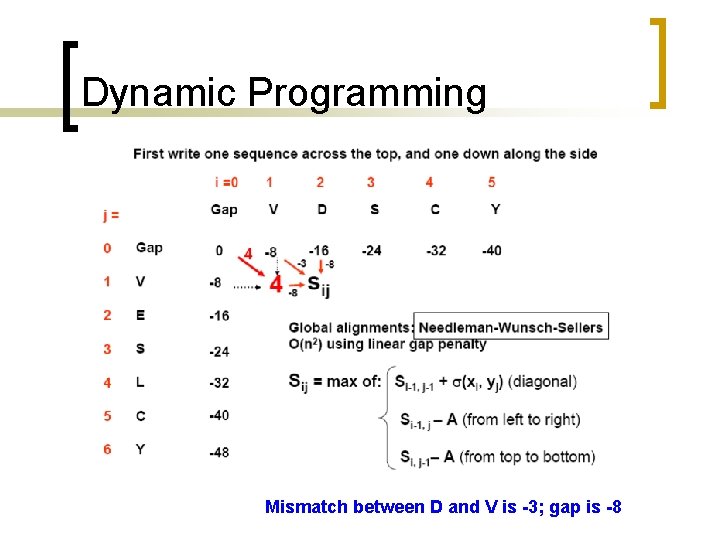
Dynamic Programming Mismatch between D and V is -3; gap is -8

Dynamic Programming http: //ocw. mit. edu/Ocw. Web/Biology/7 -91 JSpring 2004/Course. Home/

Dynamic Programming http: //ocw. mit. edu/Ocw. Web/Biology/7 -91 JSpring 2004/Course. Home/

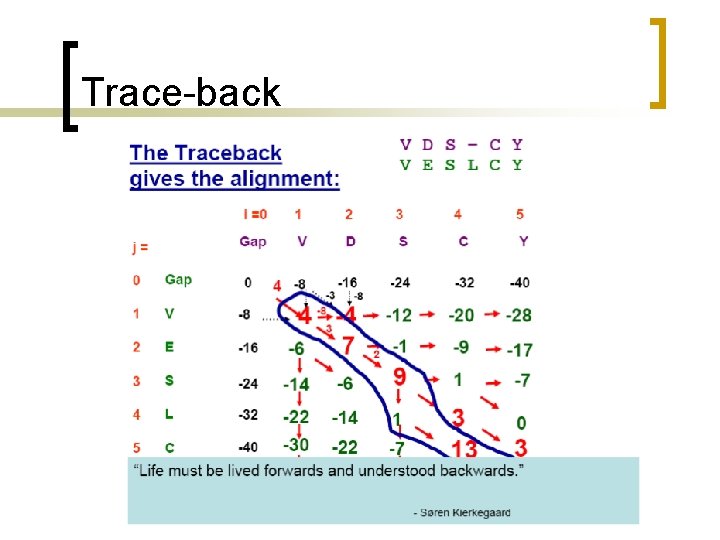
Trace-back

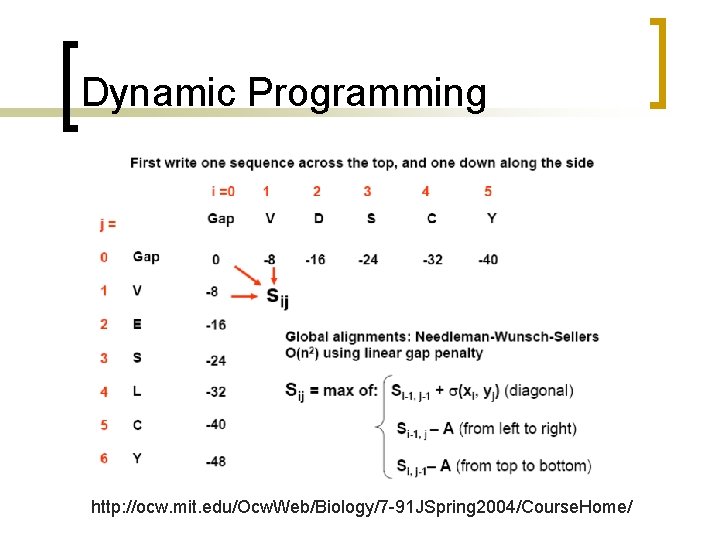
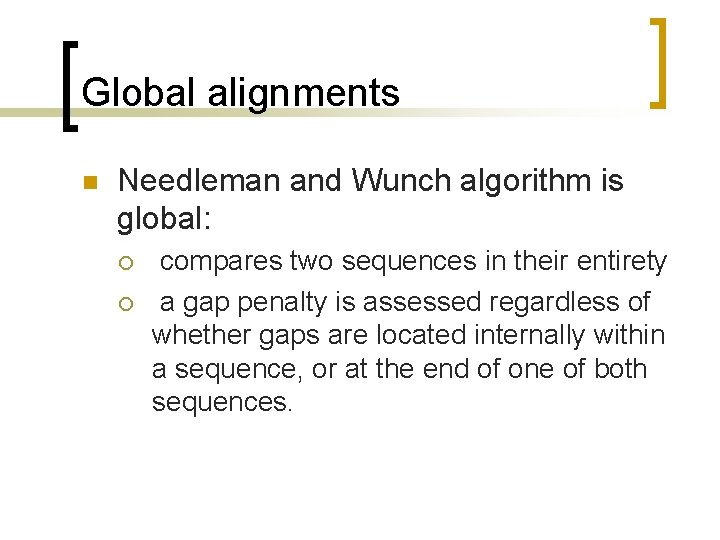
Global alignments n Needleman and Wunch algorithm is global: ¡ ¡ compares two sequences in their entirety a gap penalty is assessed regardless of whether gaps are located internally within a sequence, or at the end of one of both sequences.

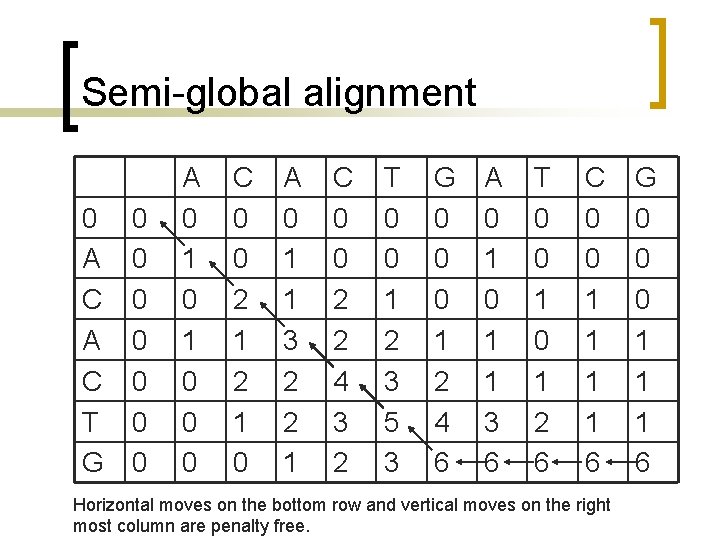
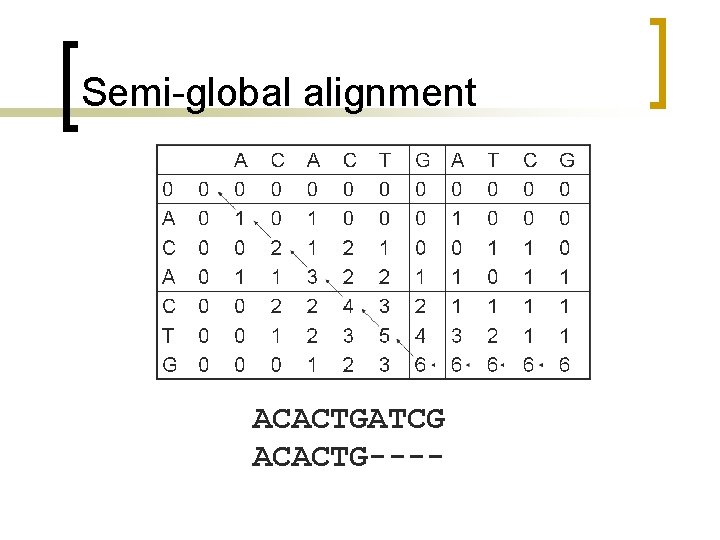
Semi-global alignment n n Terminal gaps are undermined because they are generally due to incomplete data and have no biological significance. How is it different than the global algorithm?

Semi-global vs. global n n In global a vertical move equals to a gap in the horizontal axis while a move to left equals to a gap in the vertical axis; they are both penalized. If one allows initial gaps without penalty in the sequences, should set the first row and first column of the partial scores table to 0.

Semi-global alignment 0 A C T G 0 0 0 0 A 0 1 0 0 0 C 0 0 2 1 0 A 0 1 1 3 2 2 1 C 0 0 2 2 4 3 2 T 0 0 1 2 3 5 3 G 0 0 0 1 2 4 6 A 0 1 1 3 6 T 0 0 1 2 6 C 0 0 1 1 6 Horizontal moves on the bottom row and vertical moves on the right most column are penalty free. G 0 0 0 1 1 1 6

Semi-global alignment ACACTGATCG ACACTG----

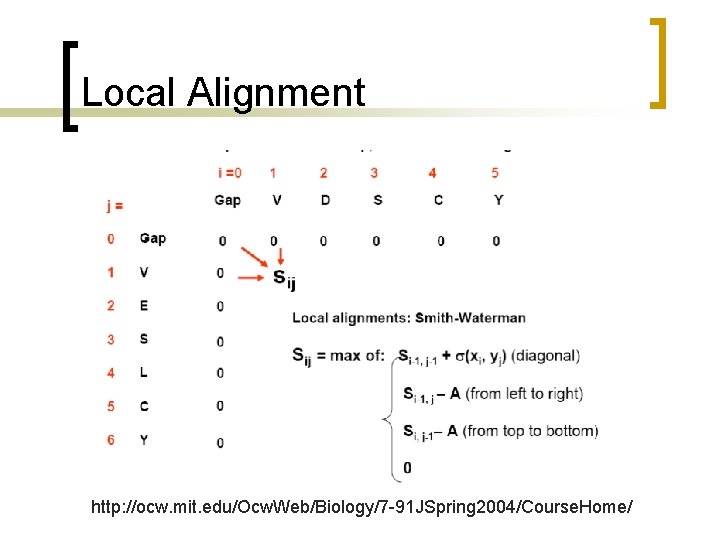
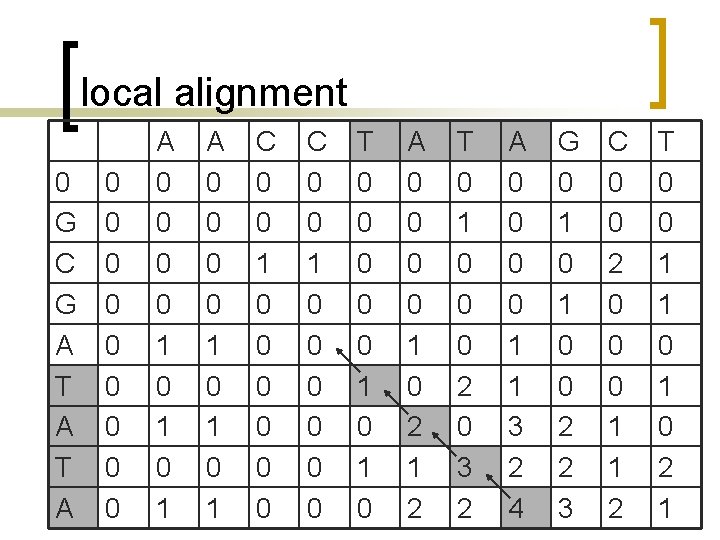
Local alignment n n n Smith-Waterman Algorithm If you have a long sequence, and want to find any subsequences that are similar to any part of yeast genome. Local alignment finds the best matching subsequences within two search sequences.

Modify global alignment algorithm n Smith-Waterman Algorithm ¡ Place a zero in any position in the table if all of the other methods result in scores lower than zero.

Local Alignment http: //ocw. mit. edu/Ocw. Web/Biology/7 -91 JSpring 2004/Course. Home/

local alignment 0 G C G A T A 0 0 0 0 0 A 0 0 0 0 1 0 1 0 1 C 0 0 1 0 0 0 T 0 0 0 1 0 A 0 0 1 0 2 1 2 T 0 1 0 0 0 2 0 3 2 A 0 0 1 1 3 2 4 G 0 1 0 0 2 2 3 C 0 0 2 0 0 0 1 1 2 T 0 0 1 1 0 2 1

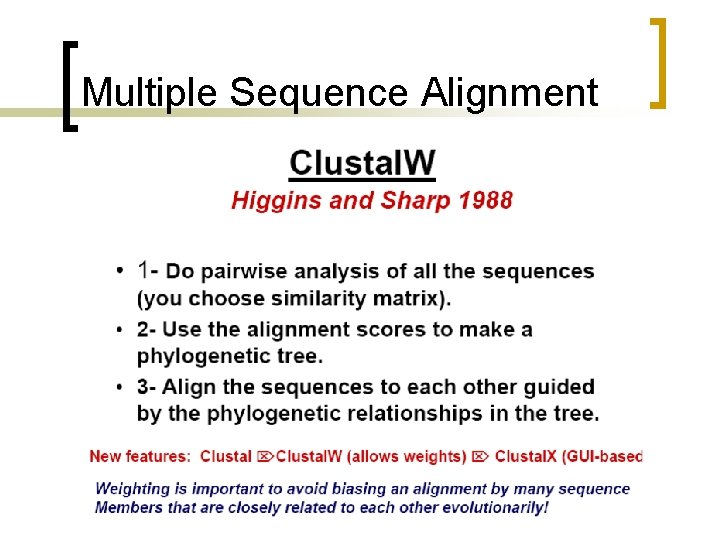
Multiple Sequence Alignment

GAPS http: //ocw. mit. edu/Ocw. Web/Biology/7 -91 JSpring 2004/Course. Home/

Use align 1 to align: enter x and y in the command line (>>) >> x='ggagaggat' x= ggagaggat >> y='ccagacct' y= ccagacct >> align 1

Use align 1 to align: enter x and y in the command line (>>) >>align 1 xalign = ggagaggat yalign = ccagacc-t ggagaggat ccagacc-t

Use alignloc 1 to align: type in alignloc 1 at the >> >>alignloc 1 xalign = aga yalign = aga aga

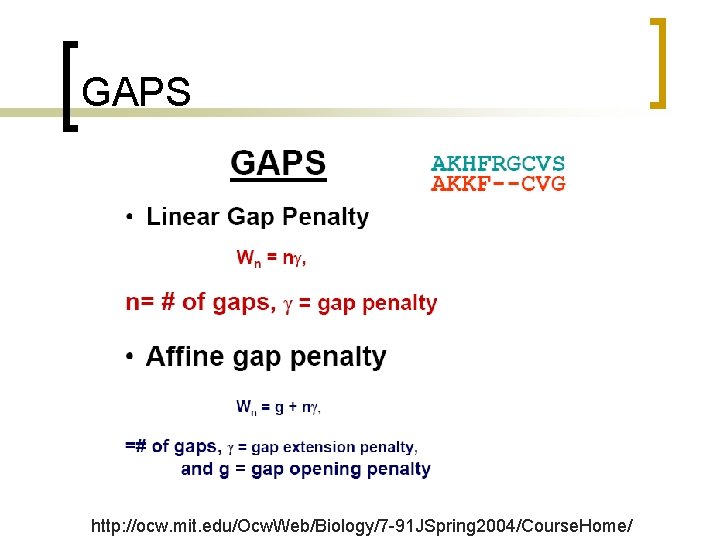
What is different? No gap penalty at the initiation %Initial Conditions for matrices F and I: for i = 2: M+1 F(i, 1) = (i-1)*0 I(i, 1) = 1 %I: vertical end for j = 2: N+1 F(1, j) = (j-1)*0 I(1, j) = 3 %I: horizontal end
![What is different Maximization Fi j Ii j maxFi1 jg Fi1 j1w Fi What is different? Maximization [F(i, j) I(i, j)] = max([F(i-1, j)+g F(i-1, j-1)+w F(i,](https://slidetodoc.com/presentation_image_h2/f8a77fe9123a7636f604200e021b66b0/image-109.jpg)
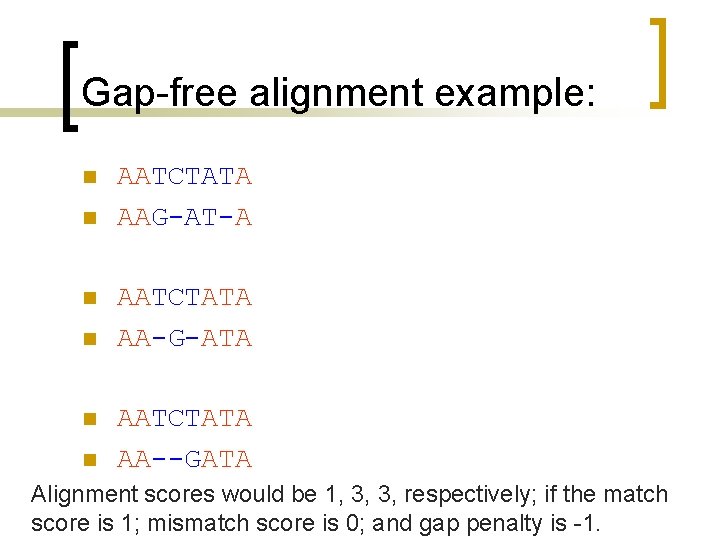
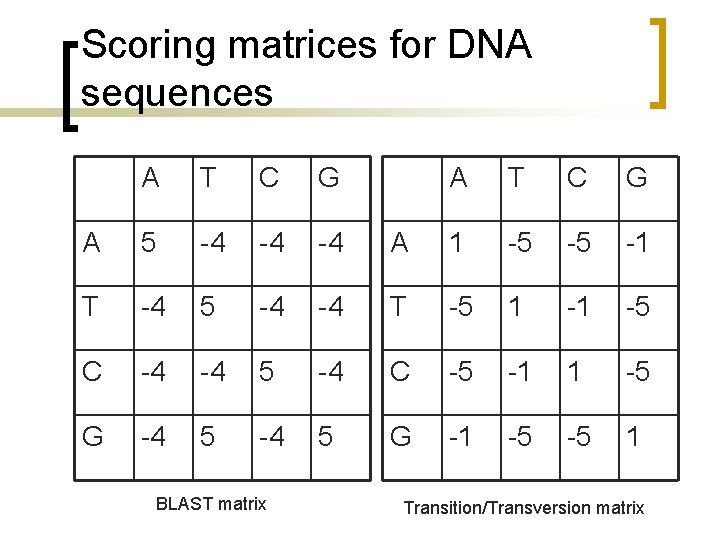
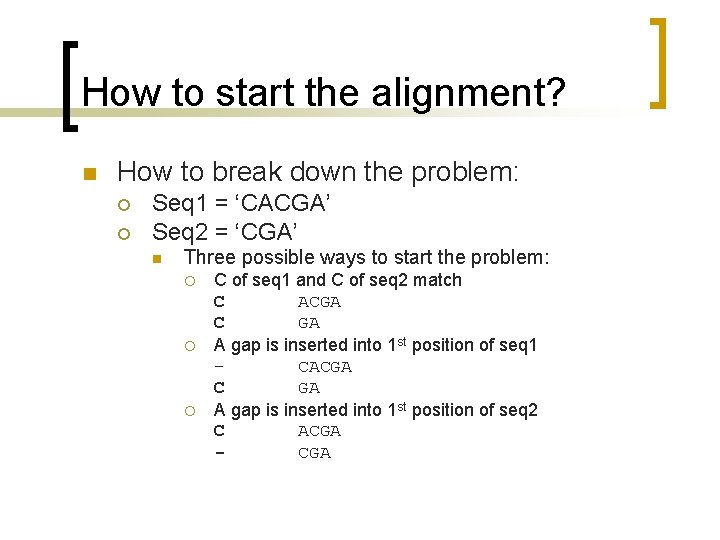
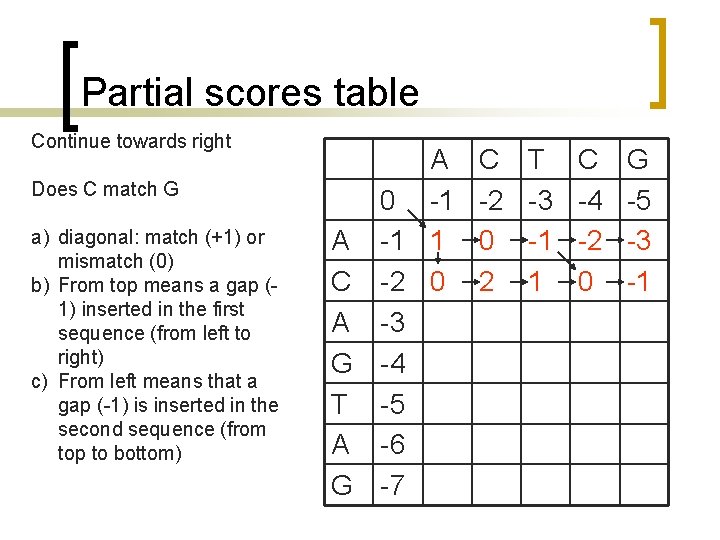
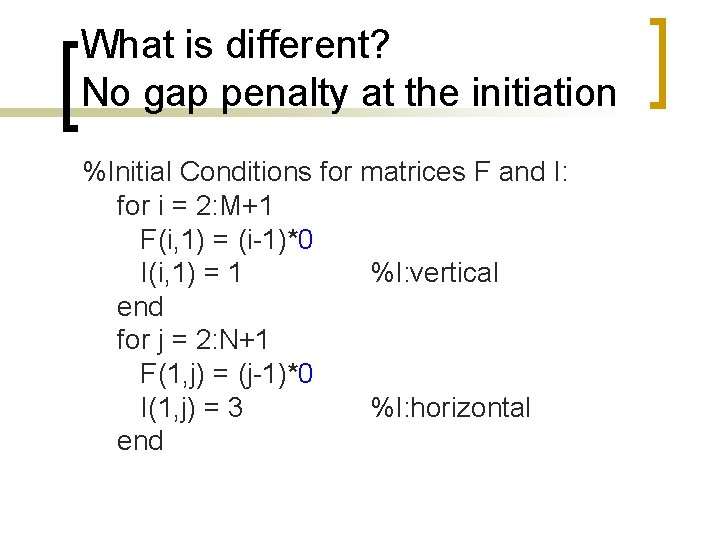
What is different? Maximization [F(i, j) I(i, j)] = max([F(i-1, j)+g F(i-1, j-1)+w F(i, j-1)+g 0])
![What is different Find the max value of the F matrix Yj fjmaxF What is different? Find the max value of the F matrix [Yj, fj]=max(F)) %](https://slidetodoc.com/presentation_image_h2/f8a77fe9123a7636f604200e021b66b0/image-110.jpg)
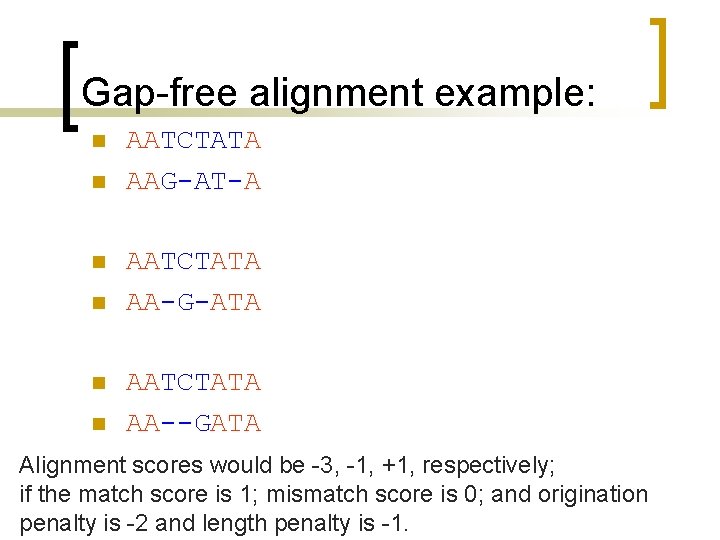
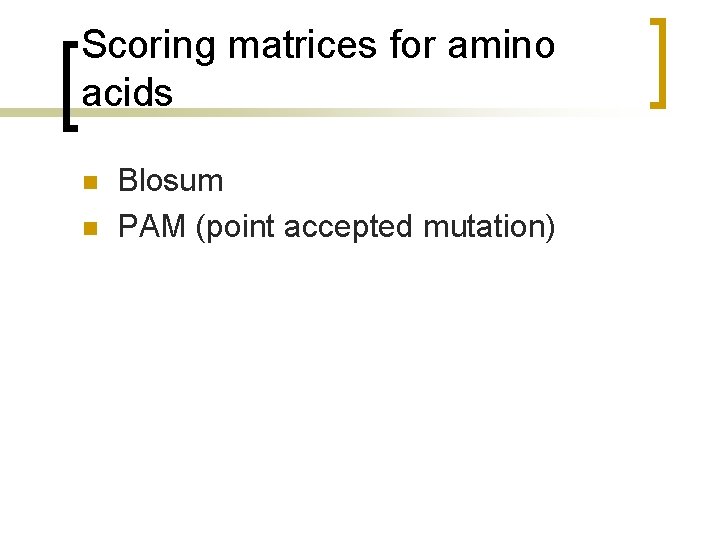
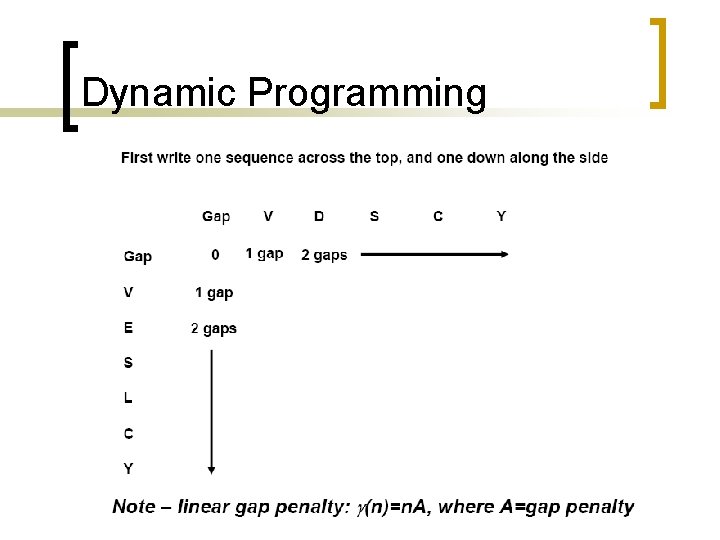
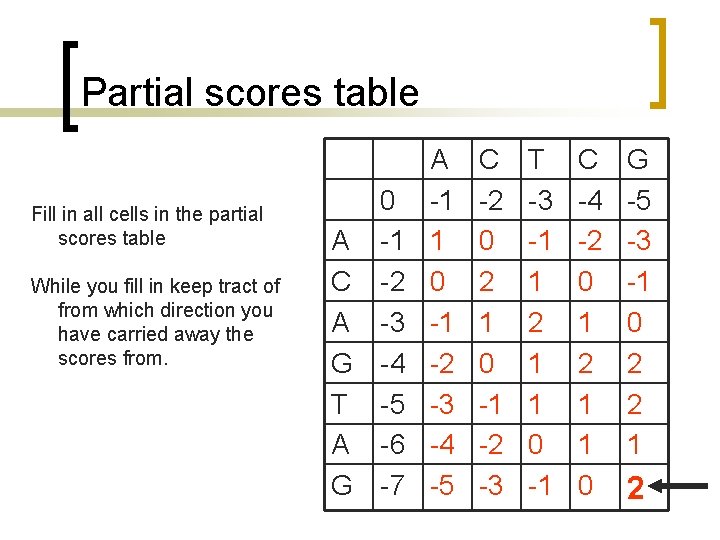
What is different? Find the max value of the F matrix [Yj, fj]=max(F)) % find column id [Yi, fi]=max(F')) % find row id i=fi(1) % set current i j=fj(1) % set current j

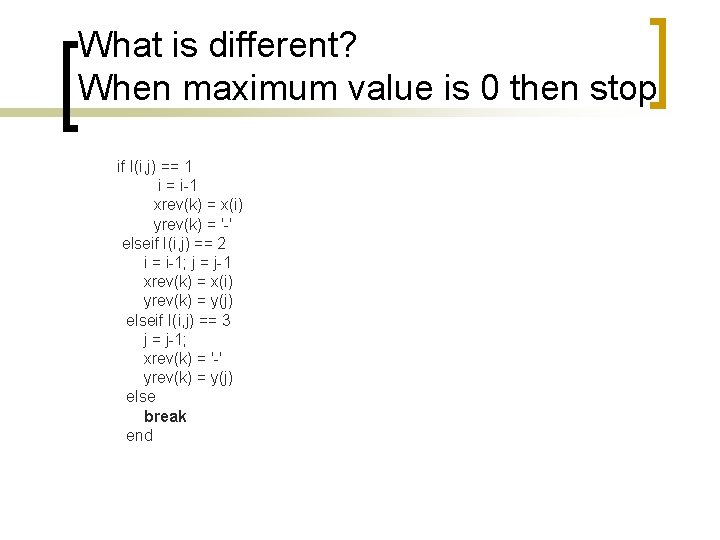
What is different? When maximum value is 0 then stop if I(i, j) == 1 i = i-1 xrev(k) = x(i) yrev(k) = '-' elseif I(i, j) == 2 i = i-1; j = j-1 xrev(k) = x(i) yrev(k) = y(j) elseif I(i, j) == 3 j = j-1; xrev(k) = '-' yrev(k) = y(j) else break end