Sequence Alignment Bioinformatics Sequence Comparison n n Problem
Sequence Alignment Bioinformatics

Sequence Comparison n n Problem: Given two sequences S & T, are S and T similar? Need to establish some notion of similarity n n n Edit distance (transforming S to T) Scoring mechanism Related Problem: Given a target sequence, obtain sequences in a database that are similar to the target

Edit Distance n n Sequences S and T are strings over an alphabet (e. g. , {a, c, t, g}) Edit operations (indels) n n n Insertion of a character Deletion of a character Example: need 3 indels to transform attc to tttac
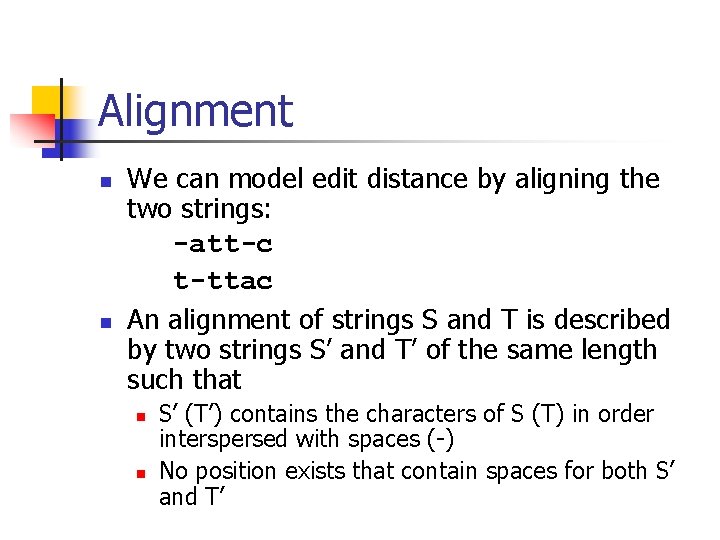
Alignment n n We can model edit distance by aligning the two strings: -att-c t-ttac An alignment of strings S and T is described by two strings S’ and T’ of the same length such that n n S’ (T’) contains the characters of S (T) in order interspersed with spaces (-) No position exists that contain spaces for both S’ and T’
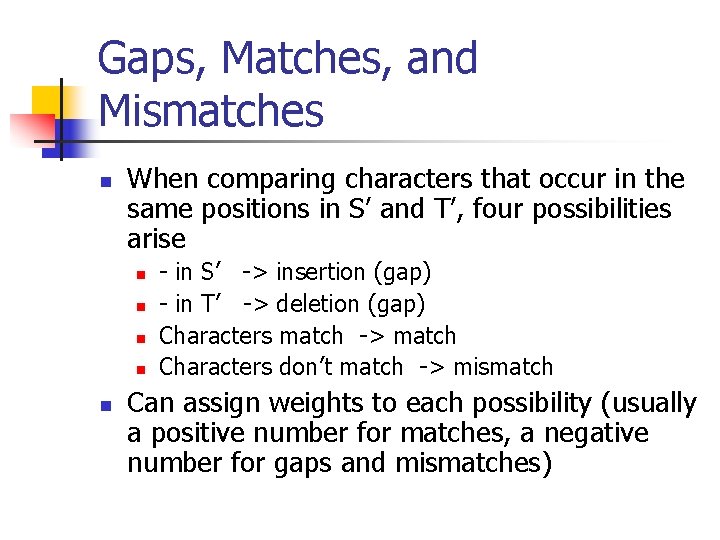
Gaps, Matches, and Mismatches n When comparing characters that occur in the same positions in S’ and T’, four possibilities arise n n n - in S’ -> insertion (gap) - in T’ -> deletion (gap) Characters match -> match Characters don’t match -> mismatch Can assign weights to each possibility (usually a positive number for matches, a negative number for gaps and mismatches)

Scoring and Optimal Alignments n Given strings S and T, and an alignment (S’, T’), a score can be computed based on pre-established weights for gaps, matches, and mismatches n n n Add all the weights for each position in S’ and T’ Note that there are many possible alignments for S and T An optimal alignment for S and T is the alignment that yields the maximum score

Problem Formulations for Sequence Comparison n n Original Formulation: Given two sequences S & T, are S and T similar? Revised Formulation: Given two sequences S & T, and weights for matches, gaps, and mismatches, determine the score of an optimal alignment of S & T
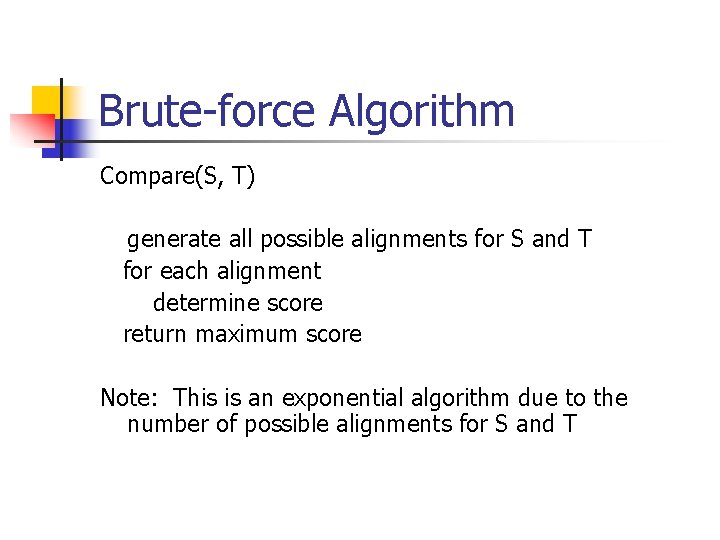
Brute-force Algorithm Compare(S, T) generate all possible alignments for S and T for each alignment determine score return maximum score Note: This is an exponential algorithm due to the number of possible alignments for S and T
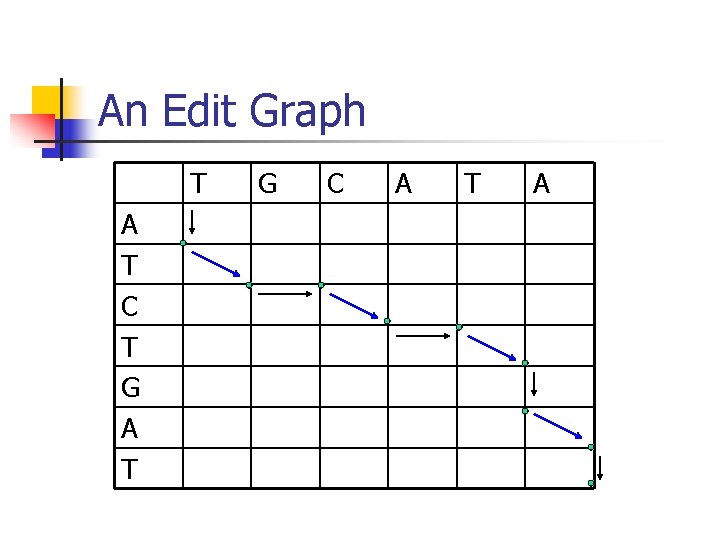
An Edit Graph T A T C T G A T G C A T A
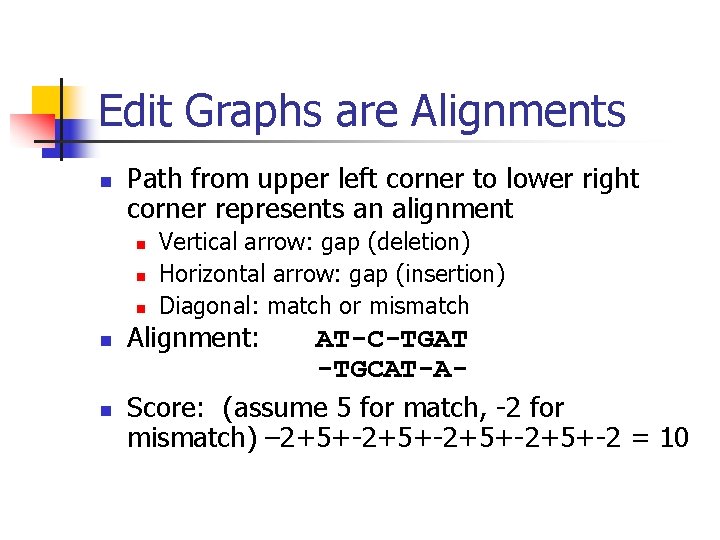
Edit Graphs are Alignments n Path from upper left corner to lower right corner represents an alignment n n n Vertical arrow: gap (deletion) Horizontal arrow: gap (insertion) Diagonal: match or mismatch Alignment: AT-C-TGAT -TGCAT-AScore: (assume 5 for match, -2 for mismatch) – 2+5+-2+5+-2 = 10
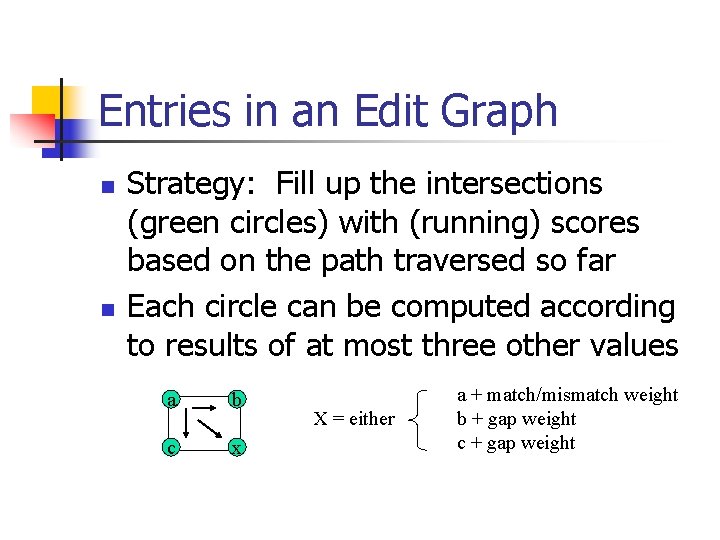
Entries in an Edit Graph n n Strategy: Fill up the intersections (green circles) with (running) scores based on the path traversed so far Each circle can be computed according to results of at most three other values a b c x X = either a + match/mismatch weight b + gap weight c + gap weight
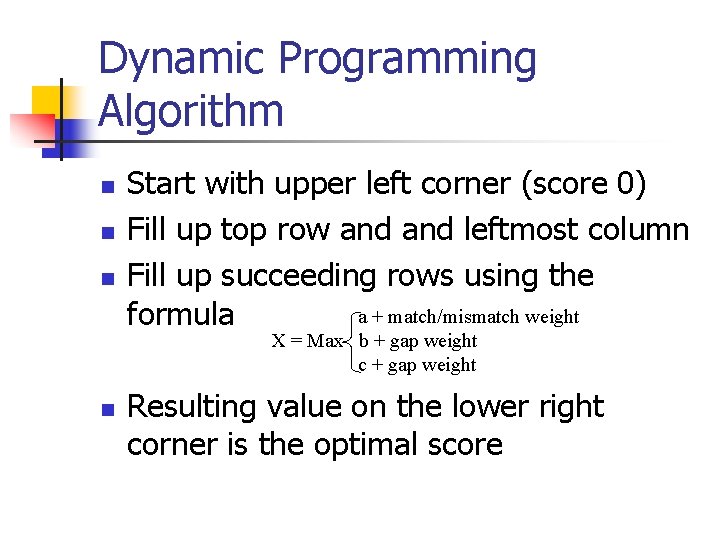
Dynamic Programming Algorithm n n n Start with upper left corner (score 0) Fill up top row and leftmost column Fill up succeeding rows using the a + match/mismatch weight formula X = Max b + gap weight c + gap weight n Resulting value on the lower right corner is the optimal score
Algorithm Analysis n n n Let N be the lengths of S and T Need to compute (N+1) entries O(N 2) algorithm
Determining the Actual Alignment n n n Need to remember which contributed to the computation of an entry (which resulting value was the maximum) Perform a back-trace from lower right corner back to the upper left corner Multiple optimal alignments possible because of ties
Other Complexity Issues n n When performing a search on a database, time complexity is dependent on the size D of the database since you run the algorithm on each sequence in the database: O(DN 2) Space requirement: an (N+1) table n Can improve to 4 N if we fill up the table according by “inverted Ls”. Topmost row and leftmost column first, then go by inner row and column, one stage at a time

Variations n n n Scoring mechanism is driven by the weights for gaps, matches and mismatches Can have different weights for starting a gap versus extending a gap (e. g. , blastp and blastn) Can have a table that allows different match/mismatch scores (e. g. , BLOSUM)
- Slides: 16