Sequence Alignment Arthur W Chou Tunghai University Fall










![Memorizing the solutions : Matrix L[ i , j ] = -1 ; // Memorizing the solutions : Matrix L[ i , j ] = -1 ; //](https://slidetodoc.com/presentation_image_h/06167f51c6640a0b7ca089ec42483fff/image-23.jpg)


![Dynamic Programming Algorithm § § L[i, j] = 1 + L[i+1, j+1] , if Dynamic Programming Algorithm § § L[i, j] = 1 + L[i+1, j+1] , if](https://slidetodoc.com/presentation_image_h/06167f51c6640a0b7ca089ec42483fff/image-26.jpg)


![Diagram Matrix S: i i+1 j j+1 s[i, j] S[i+1, j+1] Diagram Matrix S: i i+1 j j+1 s[i, j] S[i+1, j+1]](https://slidetodoc.com/presentation_image_h/06167f51c6640a0b7ca089ec42483fff/image-34.jpg)















- Slides: 64

Sequence Alignment Arthur W. Chou Tunghai University Fall 2005

Sequence Alignment Input: Output: two sequences over the same alphabet an alignment of the two sequences Example: GCGCATGGATTGAGCGA TGCGCCATTGATGACCA A possible alignment: -GCGC-ATGGATTGAGCGA TGCGCCATTGAT-GACC-A

Why align sequences? Lots of sequences don’t have known ancestry, structure, or function. A few of them do. § If they align, they are similar. § If they are similar, they might have the same ancestry, similar structure or function. § If one of them has known ancestry, structure, or function, then alignment to the others yields insight about them.

Alignments -GCGC-ATGGATTGAGCGA TGCGCCATTGAT-GACC-A Three kinds of match: Exact matches Mismatches Indels (gaps)

Choosing Alignments There are many possible alignments For example, compare: -GCGC-ATGGATTGAGCGA TGCGCCATTGAT-GACC-A to ------GCGCATGGATTGAGCGA TGCGCC----ATTGATGACCA-Which one is better?

Scoring Alignments Similar sequences evolved from a common ancestor Evolution changed the sequences from this ancestral sequence by mutations: Replacement: one letter replaced by another Deletion: deletion of a character Insertion: insertion of a character Scoring of sequence similarity should examine how many and which operations took place

Simple Scoring Rule Score each position independently: Match: Mismatch: Indel: +1 -1 -2 Score of an alignment is sum of position scores

Example -GCGC-ATGGATTGAGCGA TGCGCCATTGAT-GACC-A Score: (+1 13) + (-1 2) + (-2 4) = 3 ------GCGCATGGATTGAGCGA TGCGCC----ATTGATGACCA-Score: (+1 5) + (-1 6) + (-2 11) = -23

More General Scores The choice of +1, -1, and -2 scores is quite arbitrary Depending on the context, some changes are more plausible than others Exchange of an amino-acid by one with similar properties (size, charge, etc. ) vs. Exchange of an amino-acid by one with opposite properties Probabilistic interpretation: How likely is one alignment versus another ?

Dot Matrix Method A dot is placed at each position where two residues match. It's a visual aid. The human eye can rapidly identify similar regions in sequences. It's a good way to explore sequence organization: e. g. sequence repeats. It does not provide an alignment. This method produces dot-plots with too much noise THEFA-TCAT ||||| to be useful THEFASTCAT ð The noise can be reduced by calculating a score using a window of residues. ð The score is compared to a threshold or stringency.

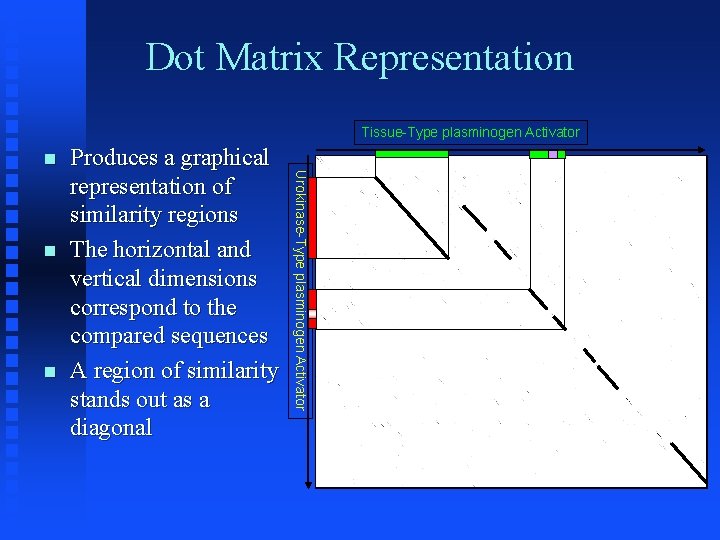
Dot Matrix Representation Tissue-Type plasminogen Activator Urokinase-Type plasminogen Activator Produces a graphical representation of similarity regions The horizontal and vertical dimensions correspond to the compared sequences A region of similarity stands out as a diagonal

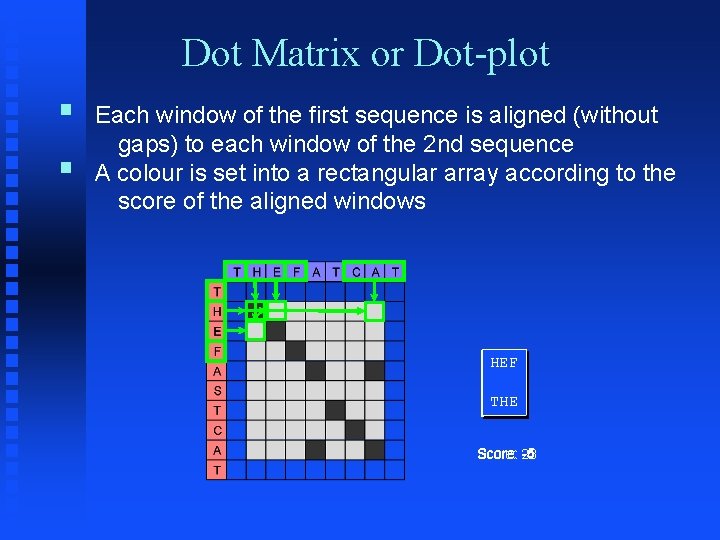
Dot Matrix or Dot-plot § Each window of the first sequence is aligned (without gaps) to each window of the 2 nd sequence § A colour is set into a rectangular array according to the score of the aligned windows HEF THE CAT ||| THE HEF Score: -5 Score: 23 Score: -4

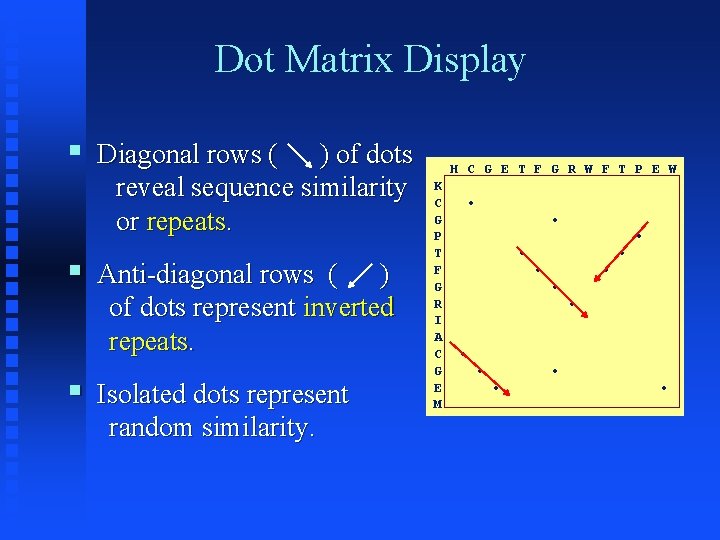
Dot Matrix Display § Diagonal rows ( ) of dots reveal sequence similarity or repeats. § Anti-diagonal rows ( ) of dots represent inverted repeats. § Isolated dots represent random similarity. H C G E T F G R W F T P E W K C • G • P • T • F • G • R • I A C • G • • E • • M

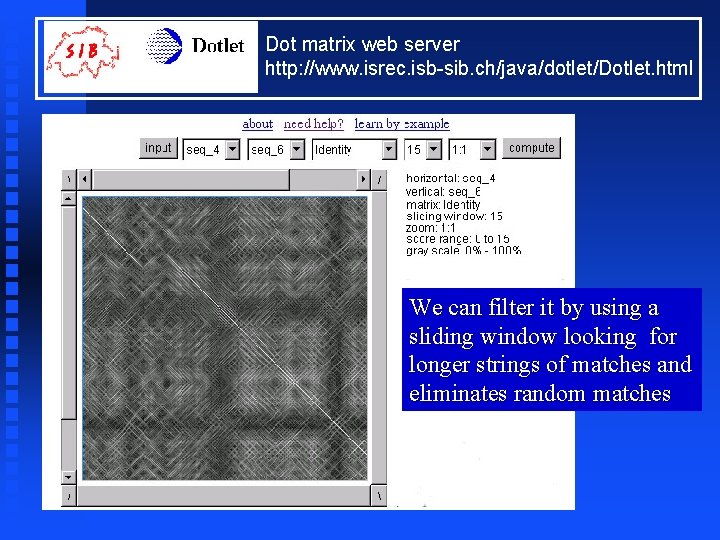
Dot matrix web server http: //www. isrec. isb-sib. ch/java/dotlet/Dotlet. html We can filter it by using a sliding window looking for longer strings of matches and eliminates random matches

Longest Common Subsequence Sequence A: Sequence B: nematode_knowledge empty_bottle n e m a t o d e _ k n o w l e d g e | | | | | e m p t y _ b o t t l e § LCS Alignment with match score 1, mismatch score 0, and gap penalty 0

What is an algorithm? A step-by-step description of the procedures to accomplish a task. Properties: 1. Determination of output for each input 2. Generality 3. Termination Criteria: 1. Correctness (proof, test, etc. ) 2. Time efficiency (no. of steps is small) 3. Space efficiency (spaced used is small)

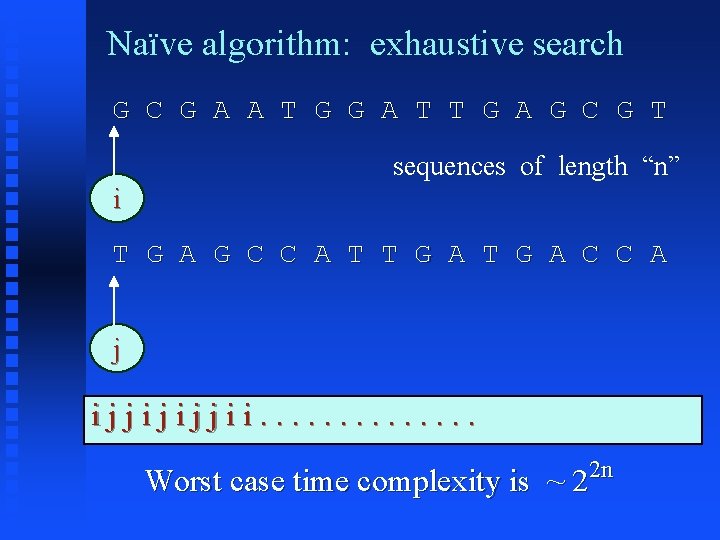
Naïve algorithm: exhaustive search G C G A A T G G A T T G A G C G T sequences of length “n” i T G A G C C A T T G A C C A j ijjijijjii. . . Worst case time complexity is ~ 2 2 n

Dynamic programming algorithms for pairwise sequence alignment Similar to Longest Common Subsequence Introduced for biological sequences by S. B. Needleman & C. D. Wunsch. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48: 443 -453 (1970)

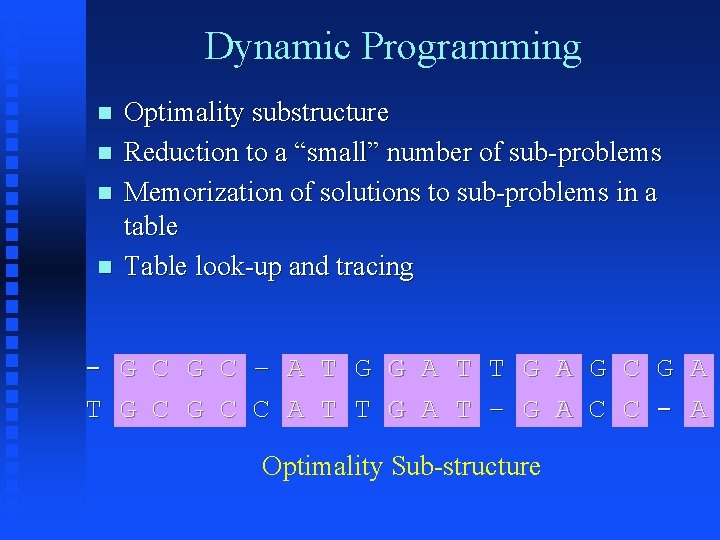
Dynamic Programming Optimality substructure Reduction to a “small” number of sub-problems Memorization of solutions to sub-problems in a table Table look-up and tracing - G C – A T G G A T T G A G C G A T G C C A T T G A T – G A C C - A Optimality Sub-structure

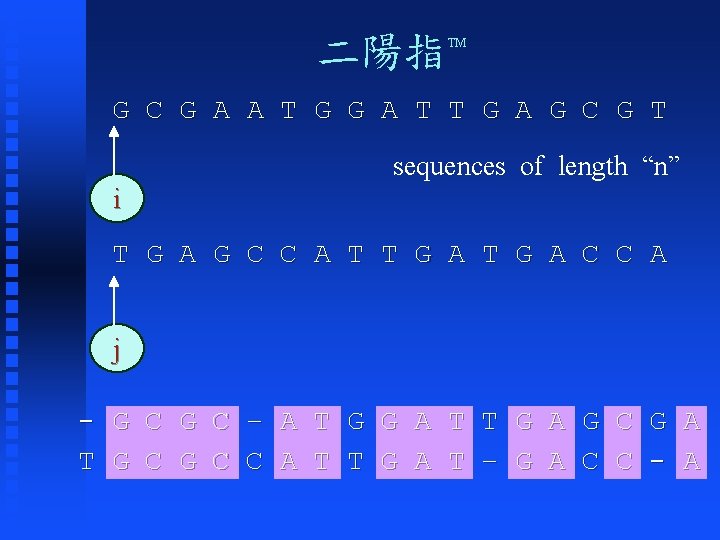
二陽指 TM G C G A A T G G A T T G A G C G T sequences of length “n” i T G A G C C A T T G A C C A j - G C – A T G G A T T G A G C G A T G C C A T T G A T – G A C C - A

Recursive LCS lcs_len( i , j ): length of LCS from i-th position onward in String A and from j-th position onward in String B int lcs_len ( i , j ) { if (A[ i ] == ‘�’ || B[ j ] == ‘�’ ) return 0 ; else if (A[ i ] == B[ j ] ) return ( 1 + lcs_len ( i+1, j+1 ) ) ; else return max ( lcs_len ( i+1, j ) , lcs_len ( i, j+1 ) ); }

Reduction to Subproblems int lcs_len ( String A , String B ) { return subproblem ( 0, 0 ); } int subproblem ( int i, int j ) { if (A[ i ] == ‘�’ || B[ j ] == ‘�’) return 0; else if ( A[ i ] == B[ j ] ) return (1 + subproblem ( i+1, j+1 )); else return max ( subproblem ( i+1, j ) , subproblem ( i, j+1 ) ); }
![Memorizing the solutions Matrix L i j 1 Memorizing the solutions : Matrix L[ i , j ] = -1 ; //](https://slidetodoc.com/presentation_image_h/06167f51c6640a0b7ca089ec42483fff/image-23.jpg)
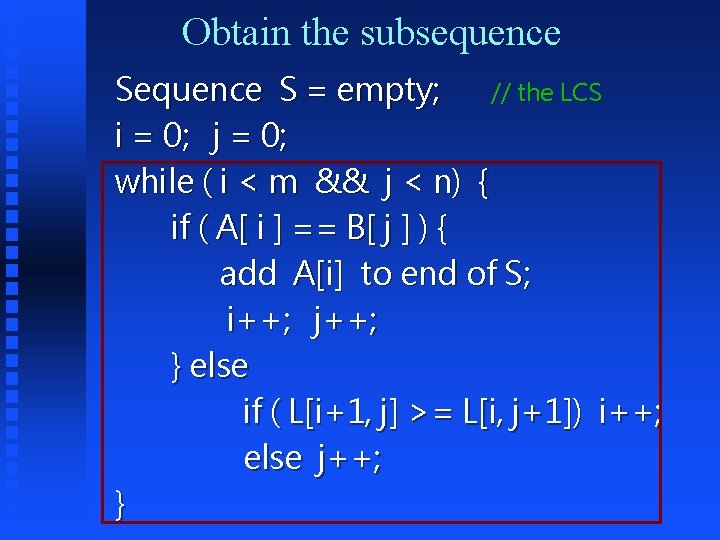
Memorizing the solutions : Matrix L[ i , j ] = -1 ; // initializing the memory device int subproblem ( int i, int j ) { if ( L[i, j] < 0 ) { if (A[ i ] == ‘�’ || B[ j ] == ‘�’) L[i , j] = 0; else if ( A[ i ] == B[ j ] ) L[i, j] = 1 + subproblem(i+1, j+1); else L[i, j] = max( subproblem(i+1, j), subproblem(i, j+1)); } return L[ i, j ] ; }

Iterative LCS: Table Look-up To get the length of LCS of A and B { first allocate storage for the matrix L; for each row i from m downto 0 for each column j from n downto 0 if (A[ i ] == ‘�’ or B[ j ] == ‘�’) L[ i, j ] = 0; else if (A[ i ] == B[ j ]) L[ i, j ] = 1 + L[i+1, j+1]; else L[ i, j ] = max(L[i+1, j], L[i, j+1]); } return L[0, 0]; }

Iterative LCS: Table Look-up int lcs_len ( String A , String B ) // find the length { // First allocate storage for the matrix L; for ( i = m ; i >= 0 ; i-- ) // A has length m+1 for ( j = n ; j >= 0 ; j-- ) { // B has length n+1 if (A[ i ] == ‘�’ || B[ j ] == ‘�’) L[ i, j ] = 0; else if (A[ i ] == B[ j ]) L[ i, j ] = 1 + L[i+1, j+1]; else L[ i, j ] = max(L[i+1, j], L[i, j+1]); } return L[0, 0]; }
![Dynamic Programming Algorithm Li j 1 Li1 j1 if Dynamic Programming Algorithm § § L[i, j] = 1 + L[i+1, j+1] , if](https://slidetodoc.com/presentation_image_h/06167f51c6640a0b7ca089ec42483fff/image-26.jpg)
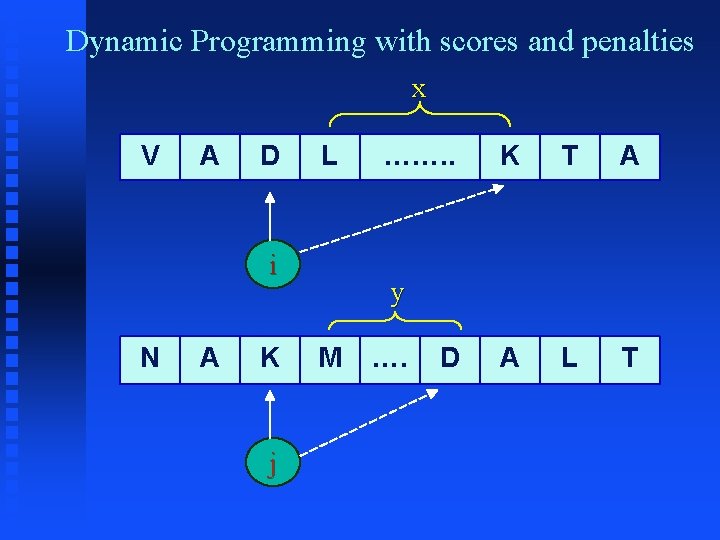
Dynamic Programming Algorithm § § L[i, j] = 1 + L[i+1, j+1] , if A[ i ] == B[ j ] ; L[i, j] = max ( L[i+1, j], L[i, j+1] ) otherwise B j j+1 i L[ i, j ] L[ i, j+1 ] i+1 L[ i+1, j ] L[i+1, j+1] A Matrix L

n e m a t o d e _ k n o w l e d g e e 7 7 6 5 5 5 4 3 3 3 2 2 2 1 1 1 0 m 6 6 6 5 5 4 4 3 3 3 2 2 1 1 0 p 5 5 5 4 4 3 3 3 2 2 1 1 0 t 5 5 5 4 4 3 3 3 2 2 1 1 0 y 4 4 4 4 4 3 3 3 2 2 1 1 0 _ 4 4 4 4 4 3 3 3 2 2 1 1 0 b 3 3 3 2 2 1 1 0 o 3 3 3 2 2 1 1 1 1 0 t 3 3 3 3 3 2 2 2 2 2 1 1 0 l 2 2 2 2 1 1 0 e 1 1 1 1 1 0 0 0 0 0

Obtain the subsequence S = empty; // the LCS i = 0; j = 0; while ( i < m && j < n) { if ( A[ i ] == B[ j ] ) { add A[i] to end of S; i++; j++; } else if ( L[i+1, j] >= L[i, j+1]) i++; else j++; }

n e m a t o d e _ k n o w l e d g e e o-o-o-o-o-o o | | | | | m o-o-o-o-o-o-o-o-o-o | | | | | | p o-o-o-o-o-o-o-o-o-o | | | | | t o-o-o-o-o-o-o-o-o-o | | | | | | y o-o-o-o-o-o-o-o-o-o | | | | | _ o-o-o-o-o-o-o-o-o-o | | | | | | | | | b o-o-o-o-o-o-o-o-o-o | | | | | o o-o-o-o o o o-o-o-o | | | | | | | t o-o-o-o-o-o-o-o-o-o | | | | | | t o-o-o-o-o-o-o-o-o-o | | | | | | l o-o-o-o-o-o-o-o | | | | | | | e o-o-o-o-o-o o | | | | | o o o o o

Dynamic Programming with scores and penalties x V A D L i N A K j ……. . K T A A L T y M …. D

Dynamic Programming with scores and penalties § from ‘i-th’ pos. in A and ‘j-th’ pos. in B onward s : score s ( A[i] , B[j] ) + S[i+1, j+1] w : penalty function S[i , j] = max best score from i, j onward max { S[i+x, j] – w( x ); gap x in sequence B } max { S[i, j+y] – w( y ); gap y in sequence A }

Algorithm for simple gap penalty If for each gap, the penalty is a fixed constant “c”, then • s(A[ i ] , B[ j ]) + S[i+1, j+1]; S[i , j] = max • S[ i+1, j ] – c ; // one gap • S[ i, j+1 ] – c ; // one gap

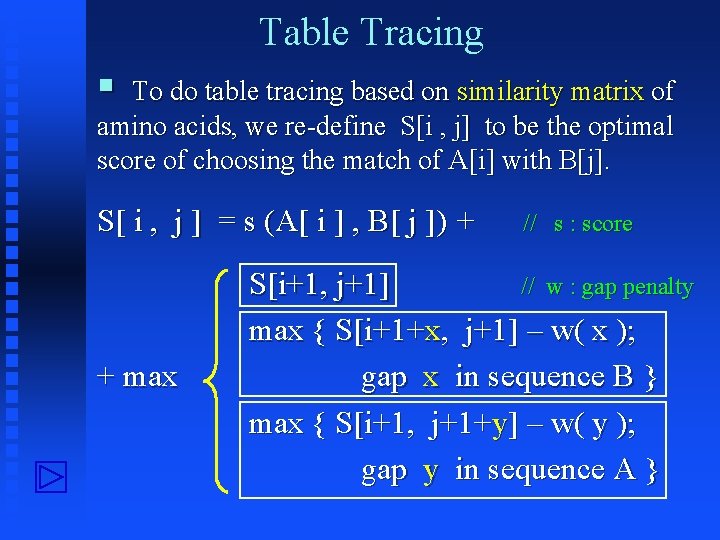
Table Tracing § To do table tracing based on similarity matrix of amino acids, we re-define S[i , j] to be the optimal score of choosing the match of A[i] with B[j]. S[ i , j ] = s (A[ i ] , B[ j ]) + + max // s : score S[i+1, j+1] // w : gap penalty max { S[i+1+x, j+1] – w( x ); gap x in sequence B } max { S[i+1, j+1+y] – w( y ); gap y in sequence A }
![Diagram Matrix S i i1 j j1 si j Si1 j1 Diagram Matrix S: i i+1 j j+1 s[i, j] S[i+1, j+1]](https://slidetodoc.com/presentation_image_h/06167f51c6640a0b7ca089ec42483fff/image-34.jpg)
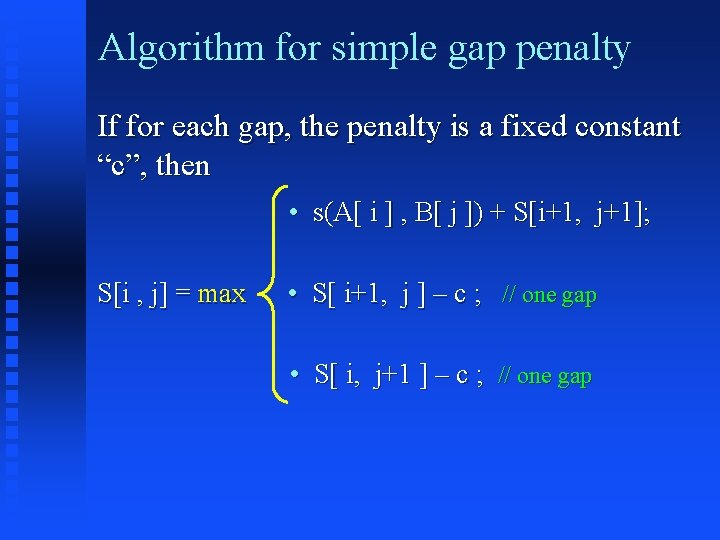
Diagram Matrix S: i i+1 j j+1 s[i, j] S[i+1, j+1]

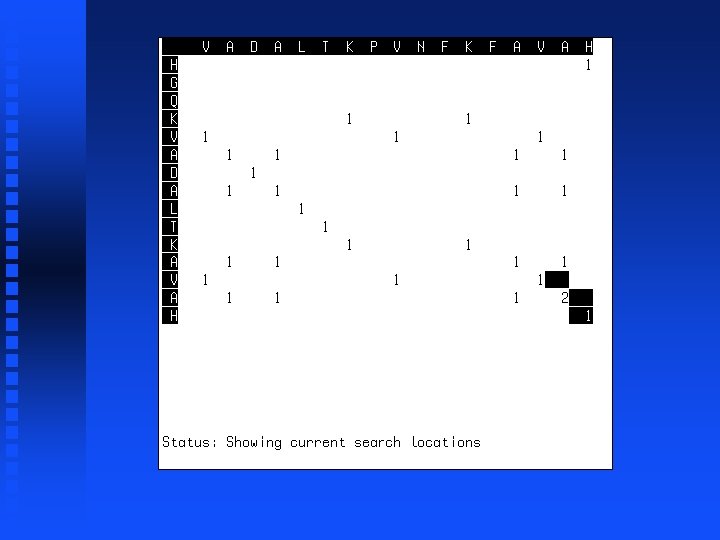
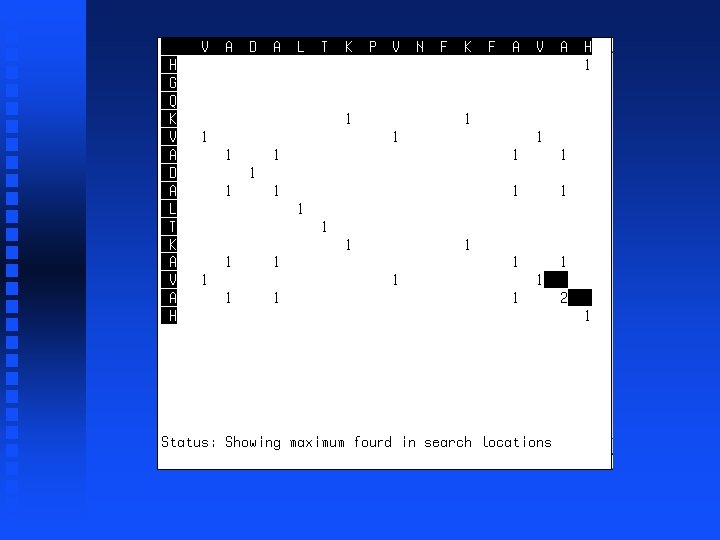
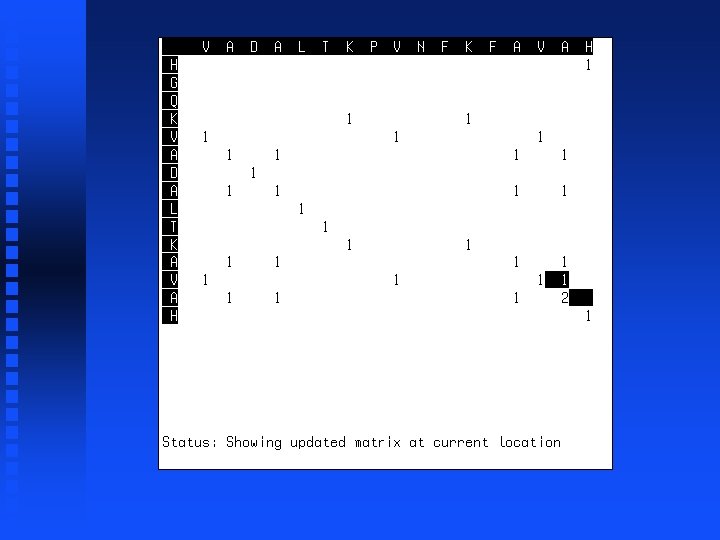
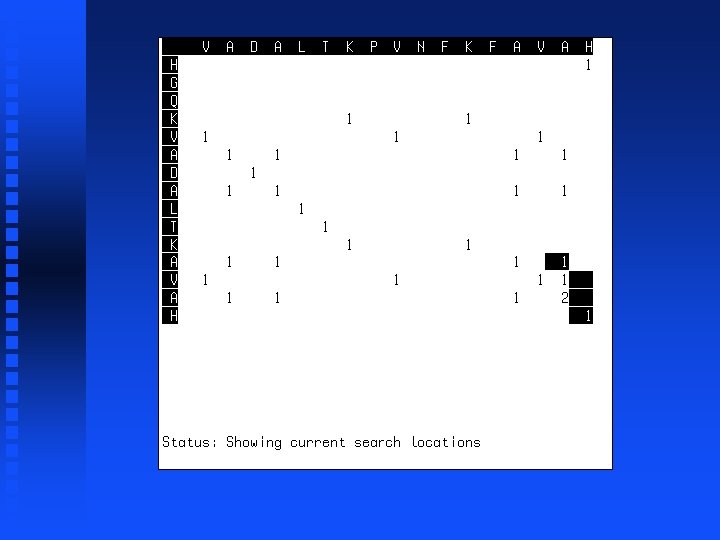
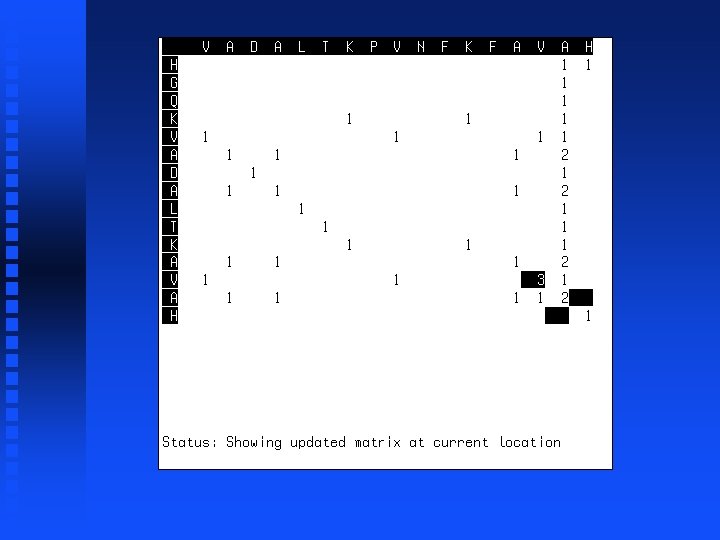
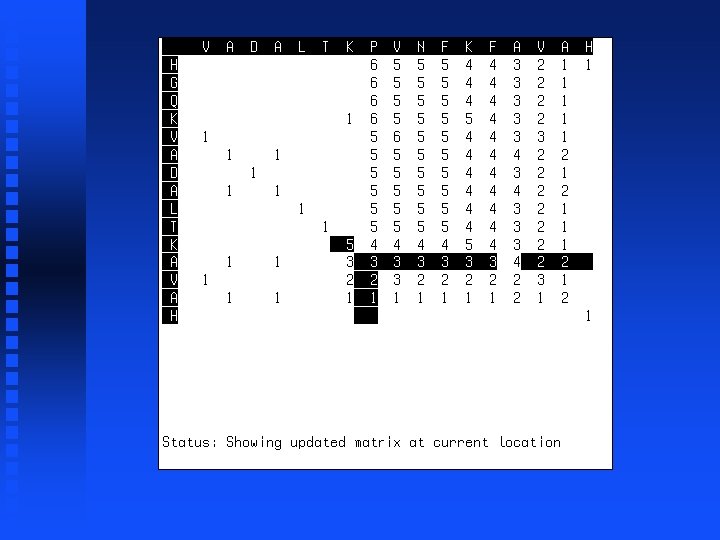
Summation operation 1. Start at lower right corner. 2. Move diagonally up one position. 3. Find largest value in either row segment starting diagonally below current position and extending to the right or column segment starting diagonally below current position and extending down. 4. Add this value to the value in the current cell. 5. Repeat steps 3 and 4 for all cells to the left in current row and all cells above in current column. 6. If we are not in the top left corner, go to step 2.
















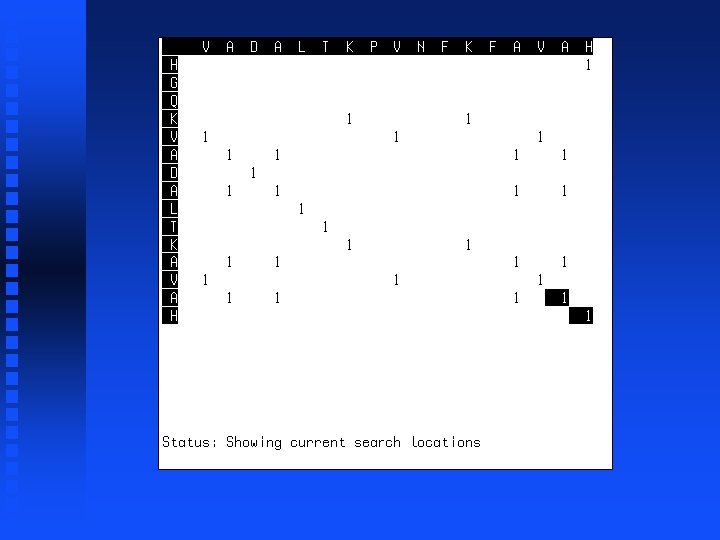
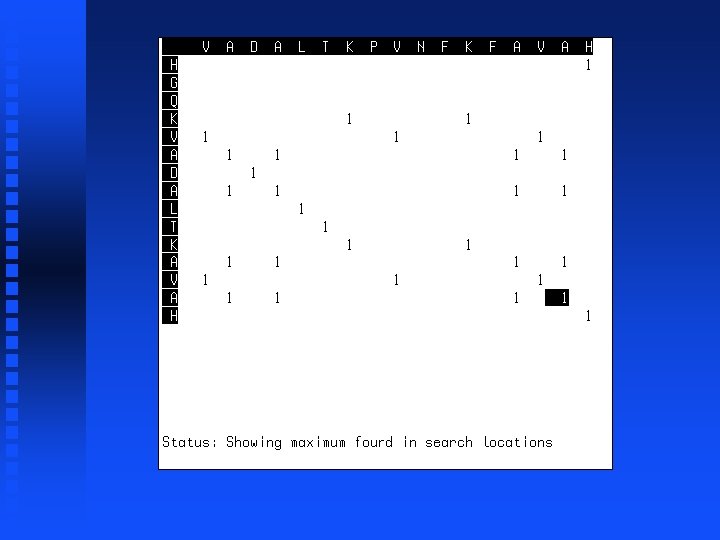
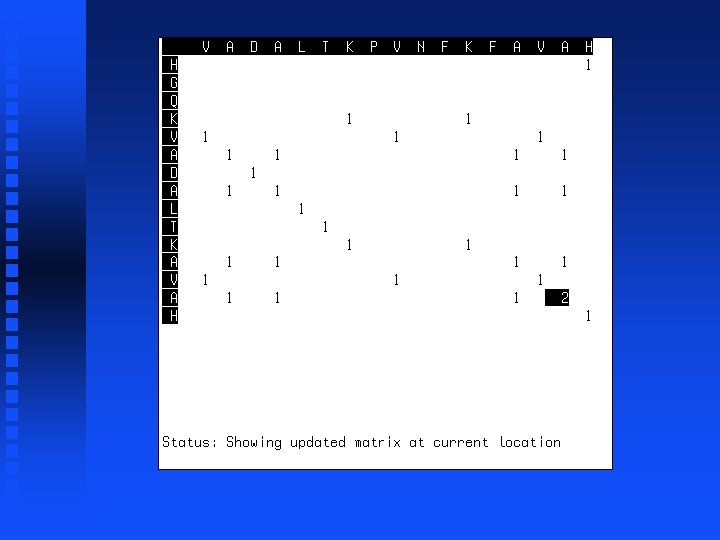
----V HGQKV


----VA HGQKVA

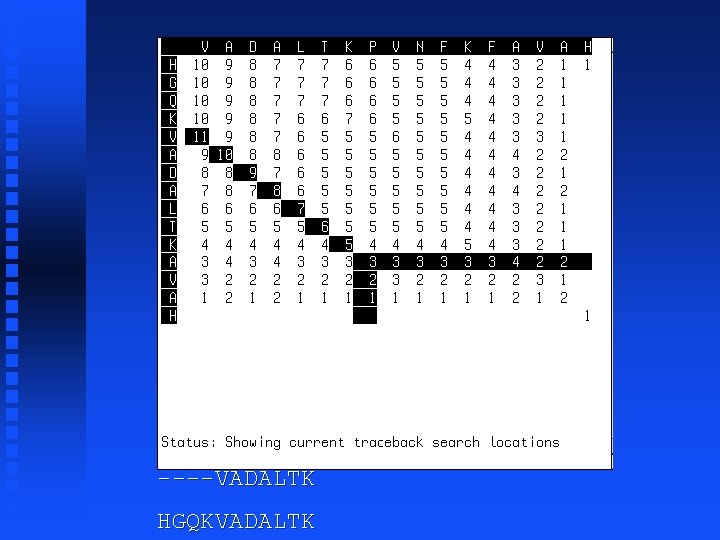
----VADALTK HGQKVADALTK

----VADALTK HGQKVADALTK

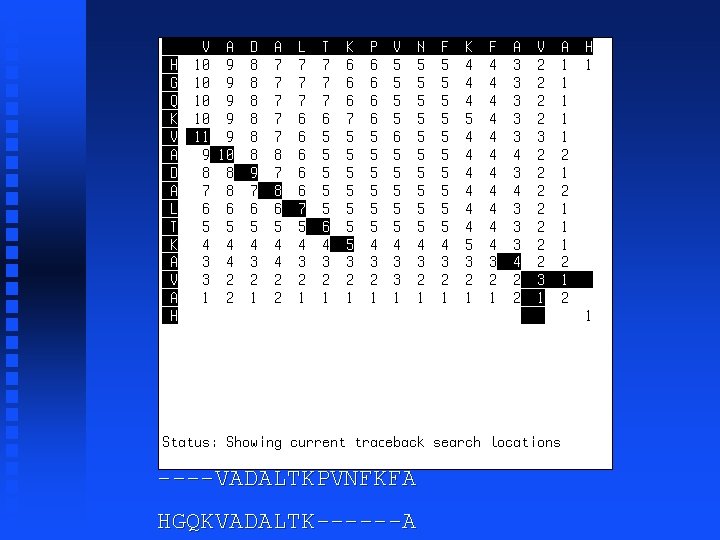
----VADALTKPVNFKFA HGQKVADALTK------A

----VADALTKPVNFKFAVAH HGQKVADALTK------AVAH

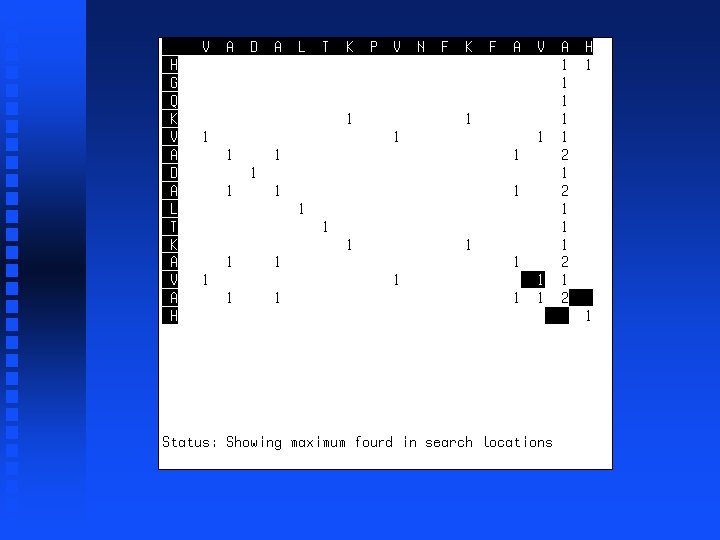
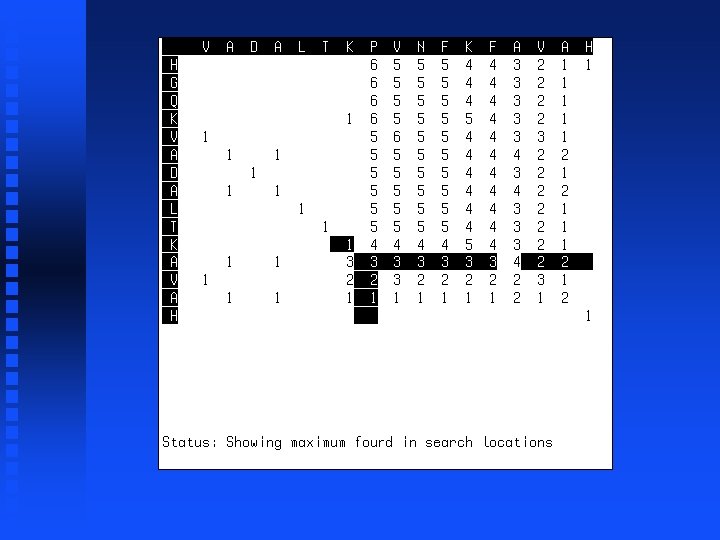
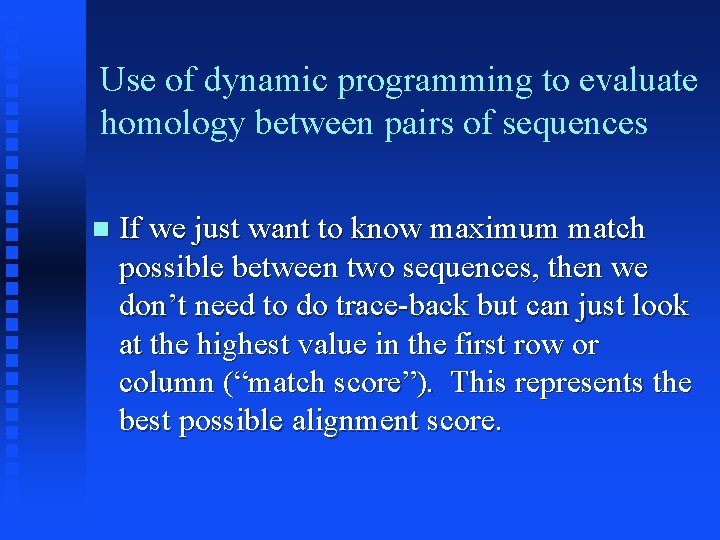
Use of dynamic programming to evaluate homology between pairs of sequences If we just want to know maximum match possible between two sequences, then we don’t need to do trace-back but can just look at the highest value in the first row or column (“match score”). This represents the best possible alignment score.

Gap penalty alternatives : constant gap penalty for gap > 1 gap penalty proportional to gap size (affine gap penalty) one penalty for starting a gap (gap opening penalty) different (lower) penalty for adding to a gap (gap extension penalty) dynamic programming algorithm can be made more efficient

Gap penalty alternatives (cont. ) gap penalty proportional to gap size and sequence for nucleic acids, can be used to mimic thermodynamics of helix formation. two kinds of gap opening penalties one for gap closed by AT, different for GC. different gap extension penalty.

End gaps Some programs treat end gaps as normal gaps and apply penalties, other programs do not apply penalties for end gaps.

End gaps (cont. ) Can determine which a program does by adding extra (unmatched) bases to the end of one sequence and seeing if match score changes. Penalties for end gaps appropriate for aligned sequences where ends "should match“. Penalties for end gaps inappropriate when surrounding sequences are expected to be different (e. g. , conserved exon surrounded by varying introns).

Global vs. Local Similarity Should result of alignment include all amino acids or proteins or just those that match? If yes, a global alignment is desired If no, a local alignment is desired Global alignment is accomplished by including negative scores for “mismatched” positions, thus scores get worse as we move away from region of match (local alignment). Instead of starting trace-back with highest value in first row or column, start with highest value in entire matrix, stop when score hits zero.

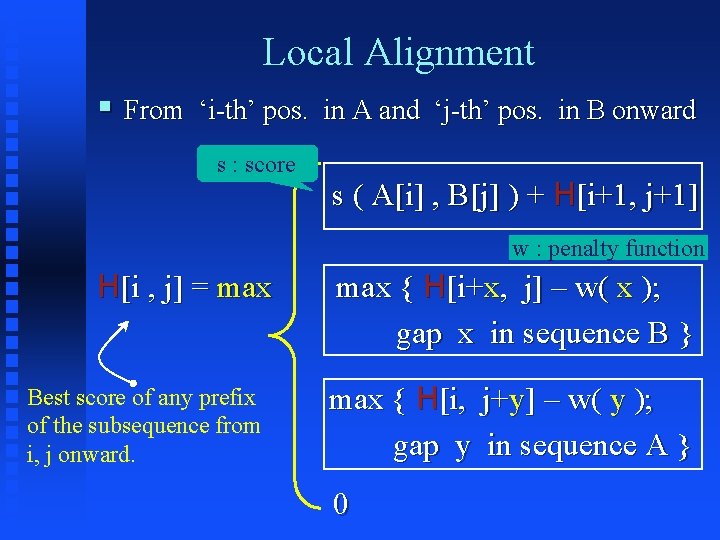
Local Alignment § From ‘i-th’ pos. in A and ‘j-th’ pos. in B onward s : score s ( A[i] , B[j] ) + H[i+1, j+1] w : penalty function H[i , j] = max Best score of any prefix of the subsequence from i, j onward. max { H[i+x, j] – w( x ); gap x in sequence B } max { H[i, j+y] – w( y ); gap y in sequence A } 0