Seq Express Introduction Features n Visualisation Tools q

- Slides: 30

Seq. Express: Introduction

Features n Visualisation Tools q q n Analysis Tools q q q n Data: gene expression, gene function and gene location. Analysis: probability models, hierarchies and clusters. Cluster analysis, refinement and validation. Using mixture modelling. Graphs and Hierarchies. Data Tools q q q Data Import/Export tools (Remote access of GEO, local access of tab separated and MAGE format). Data Integration: optional underlying data and annotation database. Data Manipulation.

Seq. Express: Visualisation Tools

Visualisations n Data Visualisation: q q n Gene Expression; Gene Variance; Gene Function/Ontology; and Chromosome Features. Analysis Visualisations: q q q Hierarchies/Graphs; Probabilistic Methods; and Cluster Comparison.

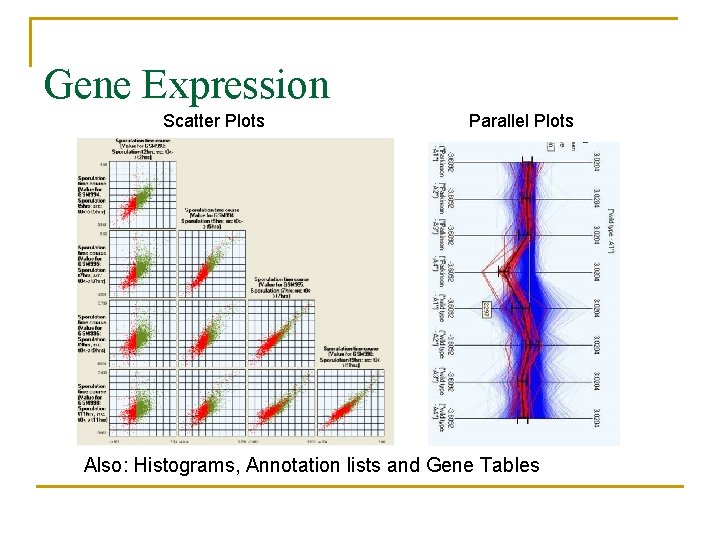
Gene Expression Scatter Plots Parallel Plots Also: Histograms, Annotation lists and Gene Tables

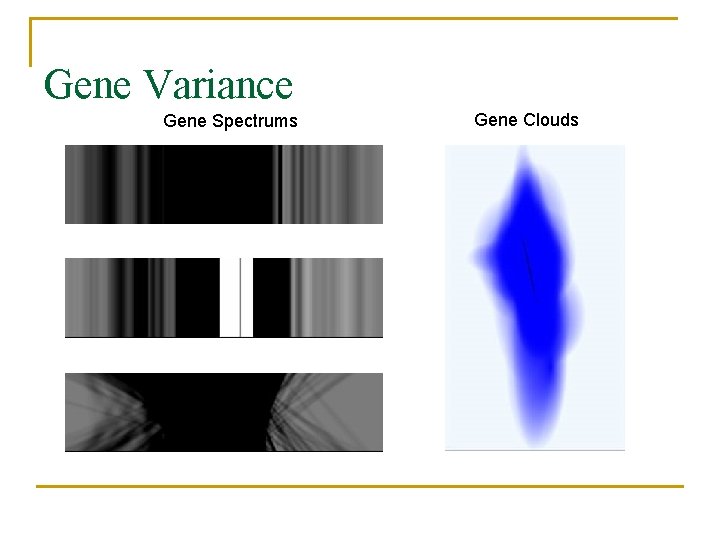
Gene Variance Gene Spectrums Gene Clouds

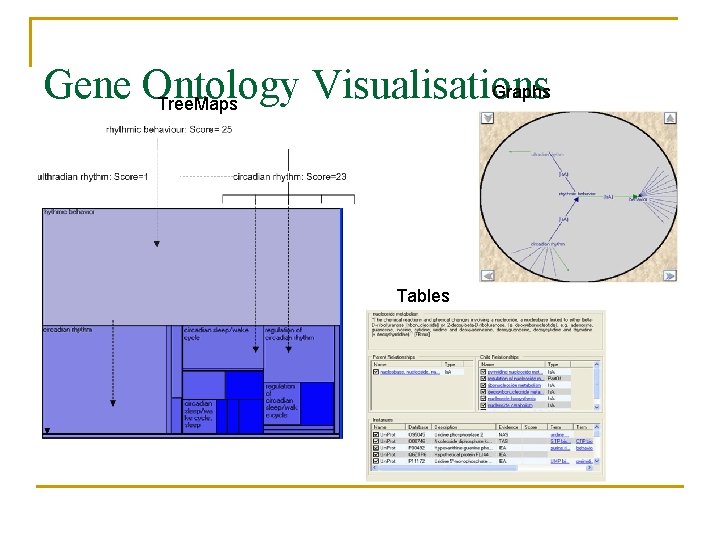
Gene Ontology Visualisations Graphs Tree. Maps Tables

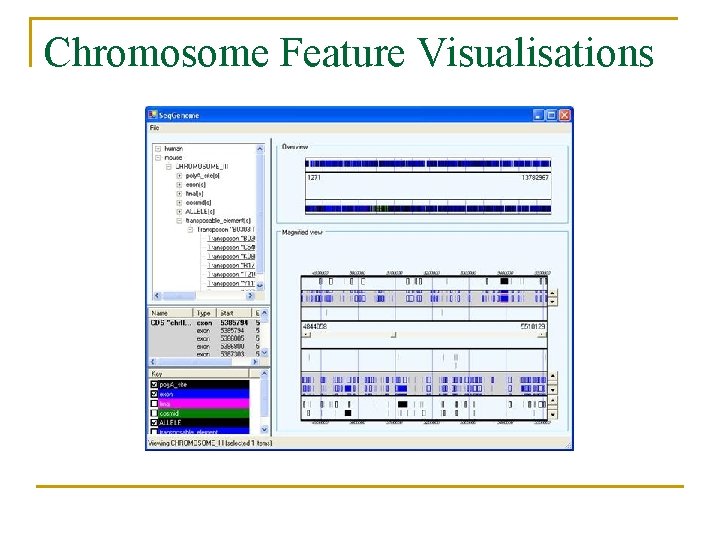
Chromosome Feature Visualisations

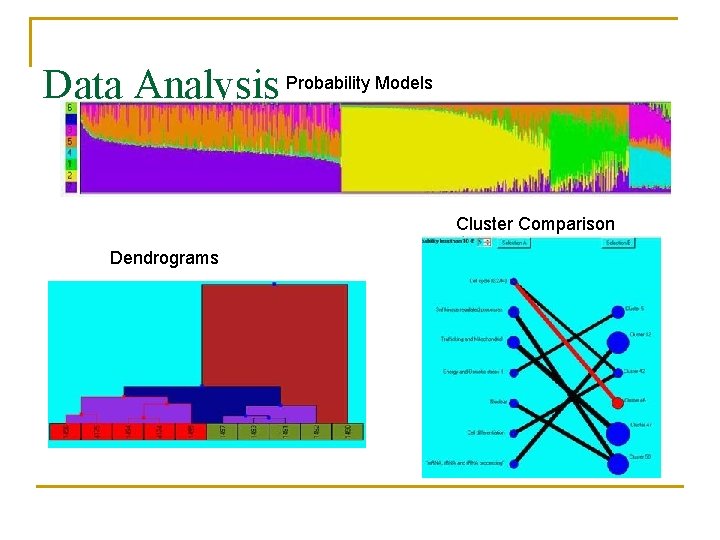
Data Analysis Probability Models Cluster Comparison Dendrograms

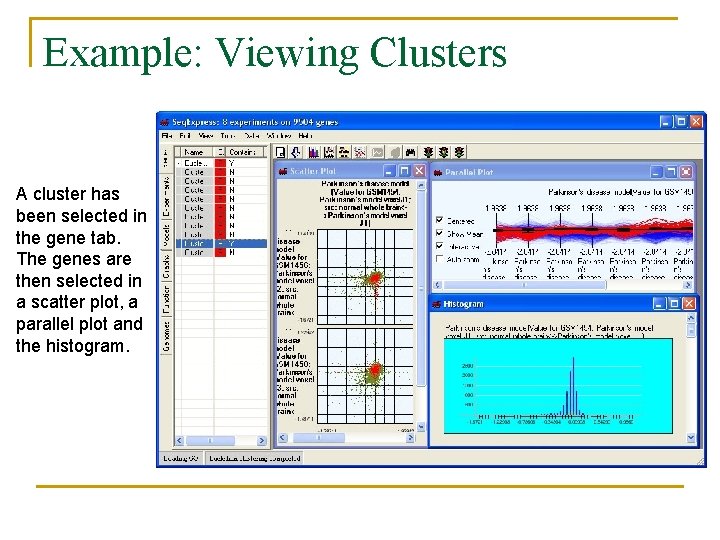
Example: Viewing Clusters A cluster has been selected in the gene tab. The genes are then selected in a scatter plot, a parallel plot and the histogram.

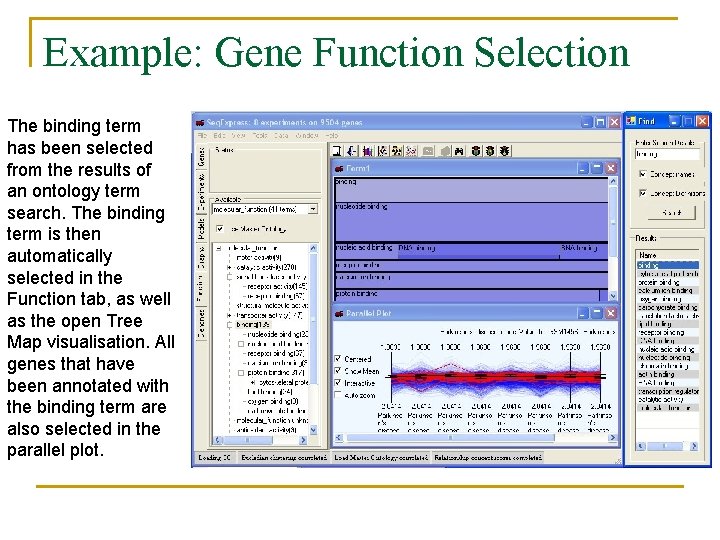
Example: Gene Function Selection The binding term has been selected from the results of an ontology term search. The binding term is then automatically selected in the Function tab, as well as the open Tree Map visualisation. All genes that have been annotated with the binding term are also selected in the parallel plot.

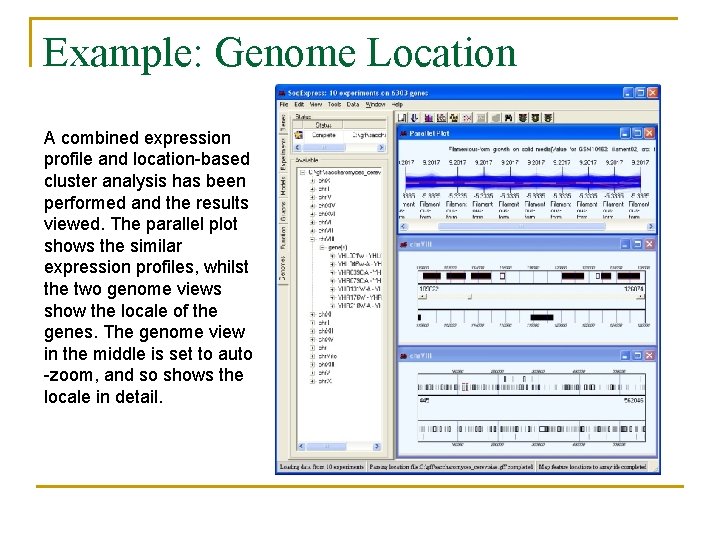
Example: Genome Location A combined expression profile and location-based cluster analysis has been performed and the results viewed. The parallel plot shows the similar expression profiles, whilst the two genome views show the locale of the genes. The genome view in the middle is set to auto -zoom, and so shows the locale in detail.

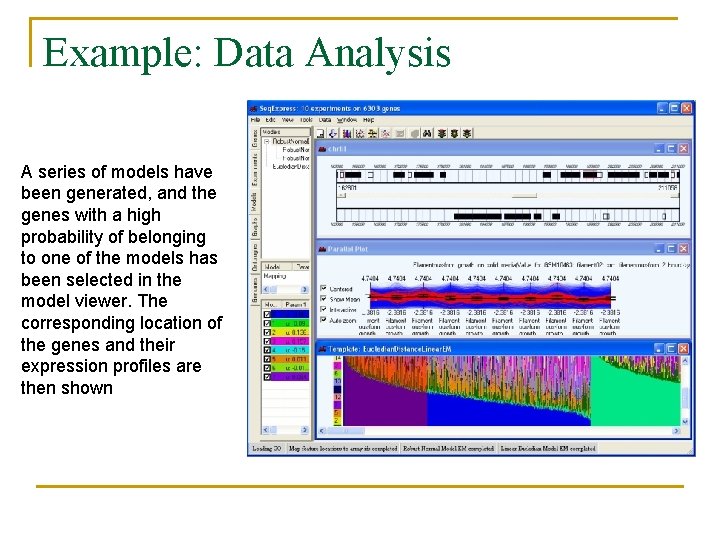
Example: Data Analysis A series of models have been generated, and the genes with a high probability of belonging to one of the models has been selected in the model viewer. The corresponding location of the genes and their expression profiles are then shown

Summary n Number of visualisations available to support variety of tasks: n n n n Expression Ontology (plus pathway and protein-protein interaction) Location Hierarchies Cluster comparison Variance Probability-theory Visualisations inter-linked

Seq. Express: Analysis Tools

Analysis Tools 1: Clusters, Hierarchies and Concepts n Clustering: q Distance based Refinement (ontology or model based). q Validation (C-Index) q n n Hierarchies: SDD*, Hierarchical Projection: q q Covariance*: eigen(covar(A)) or A=USVT Co-occurrence*: P(g, e)=P(g)ΣP(e|z)P(z|g) *Used for global/enterprise-wide information retrieval

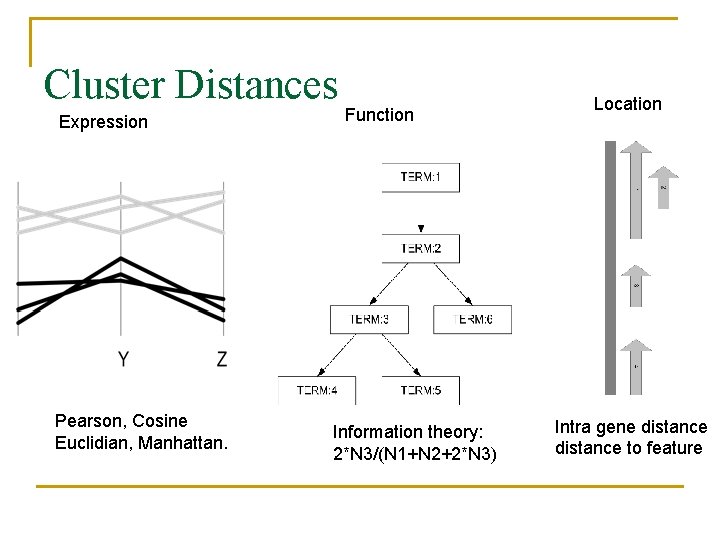
Cluster Distances Expression Pearson, Cosine Euclidian, Manhattan. Function Information theory: 2*N 3/(N 1+N 2+2*N 3) Location Intra gene distance to feature

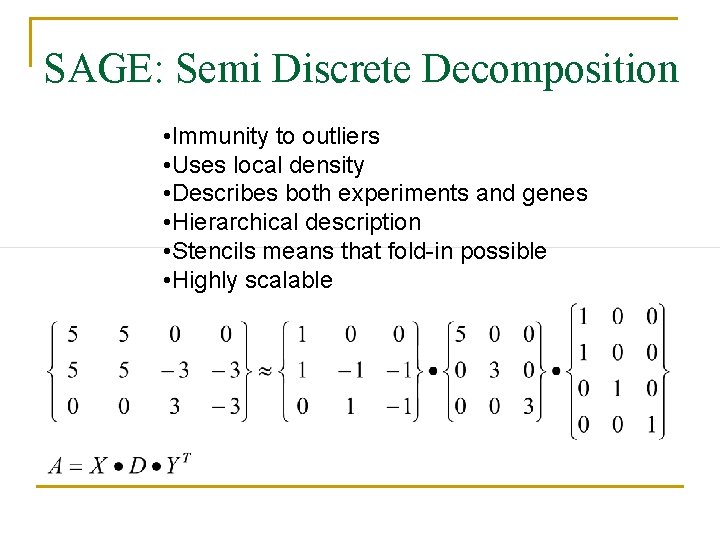
SAGE: Semi Discrete Decomposition • Immunity to outliers • Uses local density • Describes both experiments and genes • Hierarchical description • Stencils means that fold-in possible • Highly scalable

Analysis Tools 2: Models and Graphs Multi-factor analysis to identify complex features within the data (e. g. genes which have both a similar expression profile and are located on the same part of a chromosome) n n Graphs: Two factor analysis using (1)Graph Connectivity and (2) Edge Length. Models: N-factor analysis using product rule: P(A, B|C)=P(A|BC)*P(B|C).

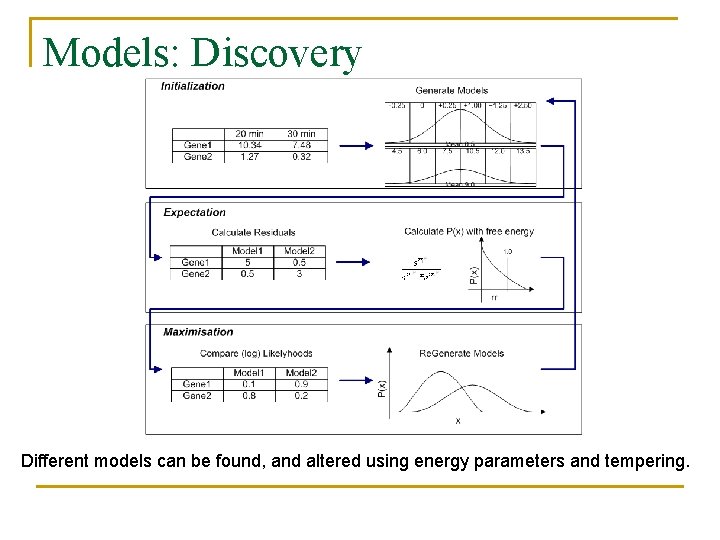
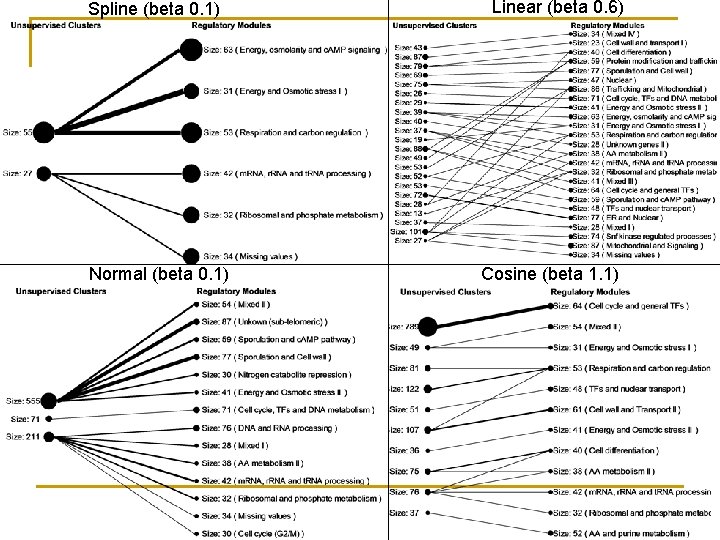
Models: Discovery Different models can be found, and altered using energy parameters and tempering.

Spline (beta 0. 1) Normal (beta 0. 1) Linear (beta 0. 6) Cosine (beta 1. 1)

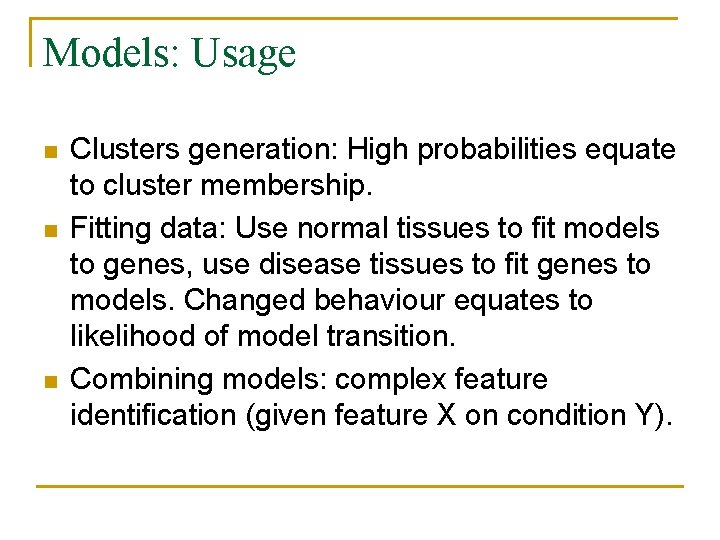
Models: Usage n n n Clusters generation: High probabilities equate to cluster membership. Fitting data: Use normal tissues to fit models to genes, use disease tissues to fit genes to models. Changed behaviour equates to likelihood of model transition. Combining models: complex feature identification (given feature X on condition Y).

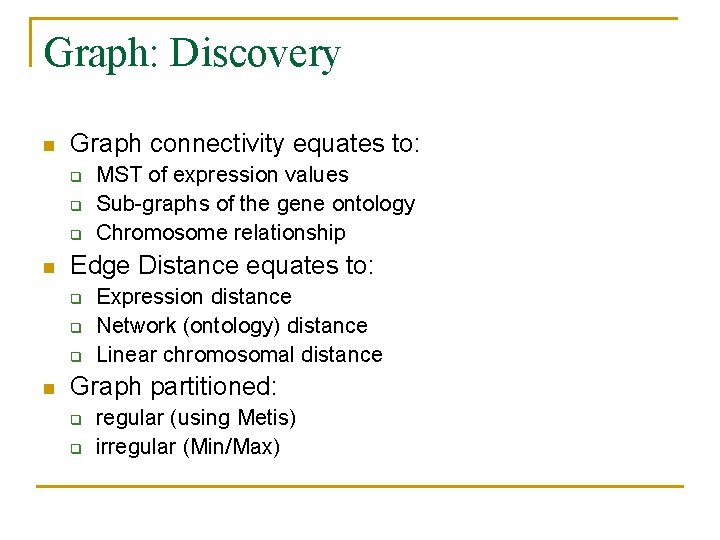
Graph: Discovery n Graph connectivity equates to: q q q n Edge Distance equates to: q q q n MST of expression values Sub-graphs of the gene ontology Chromosome relationship Expression distance Network (ontology) distance Linear chromosomal distance Graph partitioned: q q regular (using Metis) irregular (Min/Max)

Analysis: Summary n n n Desktop analysis. Number of techniques available. Techniques can be customised for different data sets (e. g. organism, array type). Borrows heavily from Information Retrieval. Probabilistic techniques show most promise.

Seq. Express: Data Tools

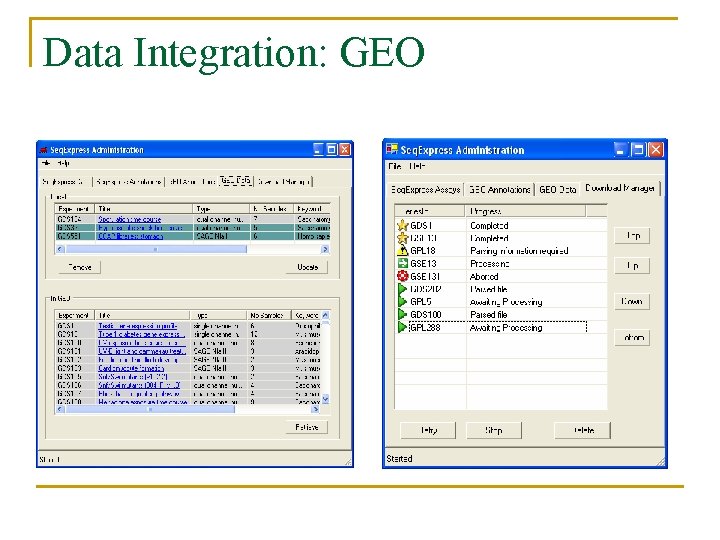
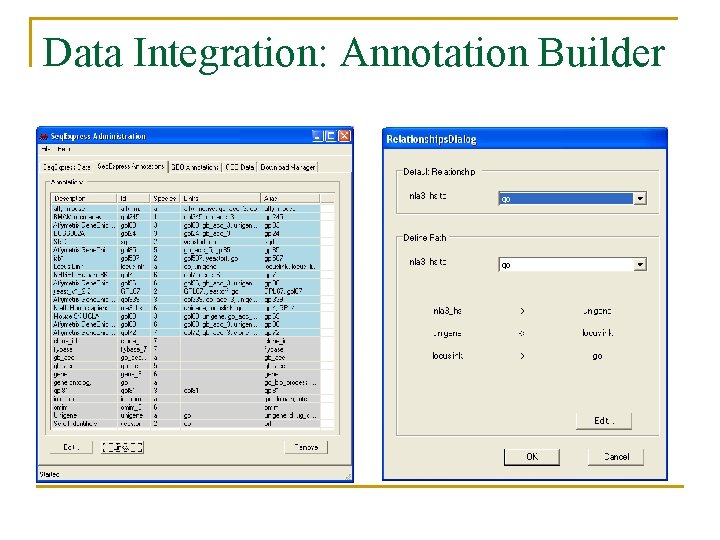
Data Analysis n Data Import/Export tools: q q q n Data Integration: data and annotation database. q n Remote access of GEO (one click access), Import tab separated and MAGE format. Export tab separated and Bioconductor format Automatic and configurable annotation mapping (e. g. SAGE tag to locuslink (entrez gene? ) to unigene) Data Manipulation: transformation, filtering and constraining

Data Integration: GEO

Data Integration: Annotation Builder

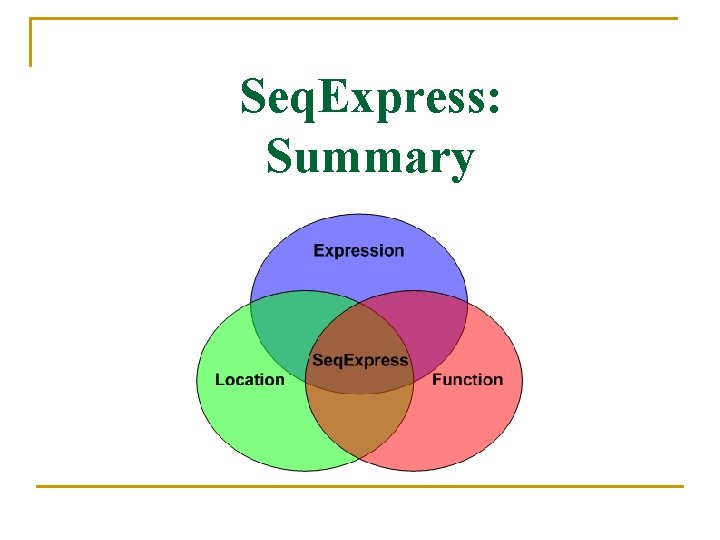
Seq. Express: Summary

Summary n n n Written in C#, is free and runs under windows. Not associated with any academic institution, funding body or commercial organisation. Development is still ongoing. Plan to develop to the Expression Application Class Specification. Looking for employment in Seattle…