Semisupervised learning for protein classification Brian R King
Semi-supervised learning for protein classification Brian R. King Chittibabu Guda, Ph. D. Department of Computer Science University at Albany, SUNY Gen*NY*sis Center for Excellence in Cancer Genomics University at Albany, SUNY 1
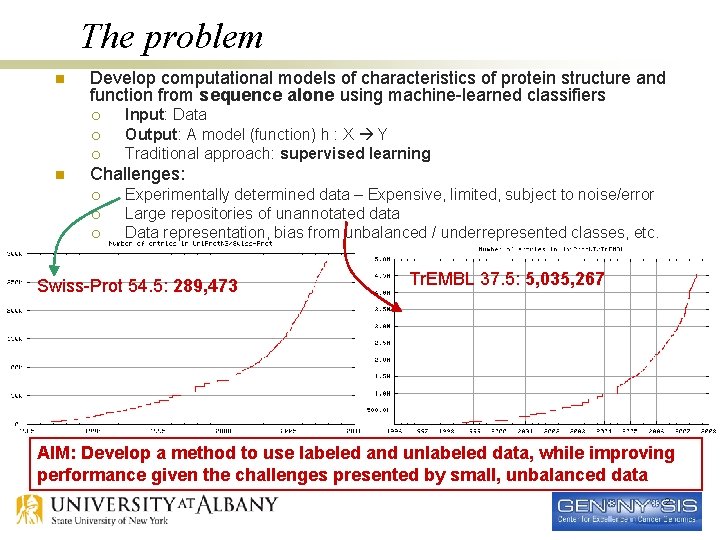
The problem n Develop computational models of characteristics of protein structure and function from sequence alone using machine-learned classifiers ¡ ¡ ¡ n Input: Data Output: A model (function) h : X Y Traditional approach: supervised learning Challenges: ¡ ¡ ¡ Experimentally determined data – Expensive, limited, subject to noise/error Large repositories of unannotated data Data representation, bias from unbalanced / underrepresented classes, etc. Swiss-Prot 54. 5: 289, 473 Tr. EMBL 37. 5: 5, 035, 267 AIM: Develop a method to use labeled and unlabeled data, while improving performance given the challenges presented by small, unbalanced data 2
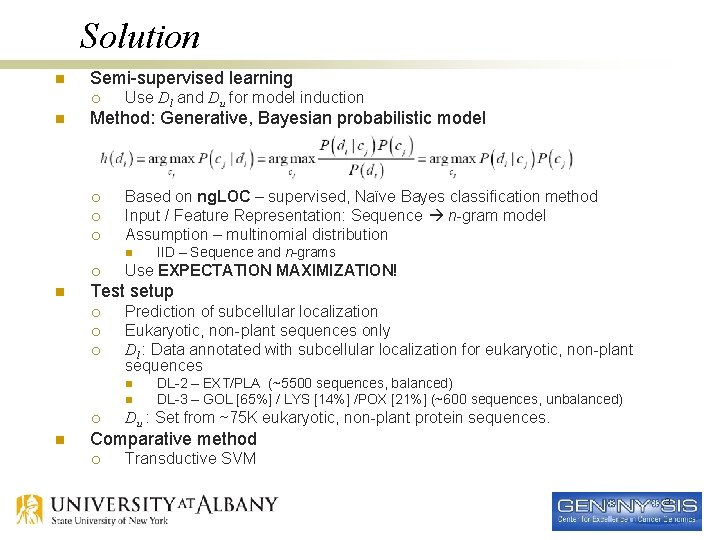
Solution n Semi-supervised learning ¡ n Use Dl and Du for model induction Method: Generative, Bayesian probabilistic model ¡ ¡ ¡ Based on ng. LOC – supervised, Naïve Bayes classification method Input / Feature Representation: Sequence n-gram model Assumption – multinomial distribution n ¡ n Use EXPECTATION MAXIMIZATION! Test setup ¡ ¡ ¡ Prediction of subcellular localization Eukaryotic, non-plant sequences only Dl : Data annotated with subcellular localization for eukaryotic, non-plant sequences n n ¡ n IID – Sequence and n-grams DL-2 – EXT/PLA (~5500 sequences, balanced) DL-3 – GOL [65%] / LYS [14%] /POX [21%] (~600 sequences, unbalanced) Du : Set from ~75 K eukaryotic, non-plant protein sequences. Comparative method ¡ Transductive SVM 3
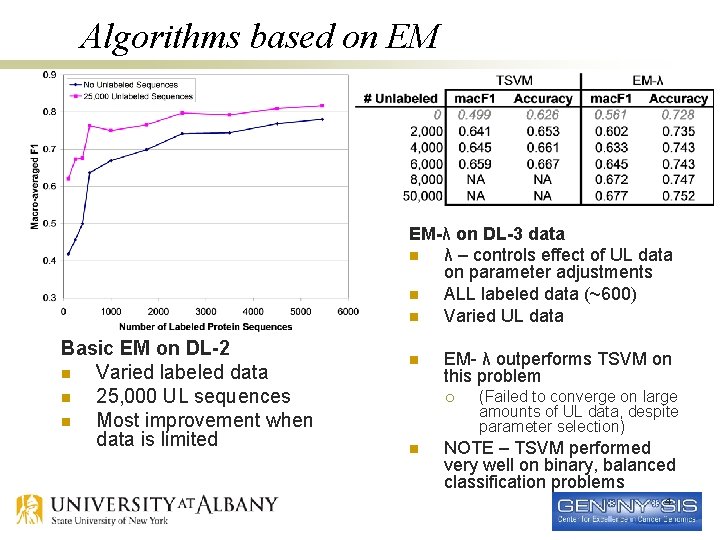
Algorithms based on EM EM-λ on DL-3 data n λ – controls effect of UL data on parameter adjustments n ALL labeled data (~600) n Varied UL data Basic EM on DL-2 n Varied labeled data n 25, 000 UL sequences n Most improvement when data is limited n EM- λ outperforms TSVM on this problem ¡ n (Failed to converge on large amounts of UL data, despite parameter selection) NOTE – TSVM performed very well on binary, balanced classification problems 4
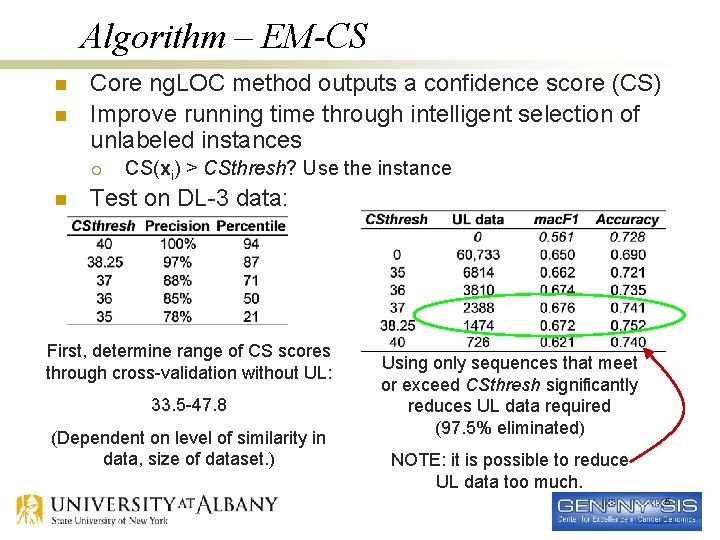
Algorithm – EM-CS n n Core ng. LOC method outputs a confidence score (CS) Improve running time through intelligent selection of unlabeled instances ¡ n CS(xi) > CSthresh? Use the instance Test on DL-3 data: First, determine range of CS scores through cross-validation without UL: 33. 5 -47. 8 (Dependent on level of similarity in data, size of dataset. ) Using only sequences that meet or exceed CSthresh significantly reduces UL data required (97. 5% eliminated) NOTE: it is possible to reduce UL data too much. 5
Conclusion n Benefits: ¡ Probabilistic n n ¡ Extract unlabeled sequences of “high-confidence” Difficult with SVM or TSVM Extraction of knowledge from model n Discriminative n-grams and anomalies ¡ n n ¡ n Time: Significantly lower than SVM and TSVM Space: Dependent on n-gram model Can use large amounts of unlabeled data Applicable toward prediction of any structural or functional characteristic Outputs a global model n ¡ Again, difficult with SVM or TSVM Computational resources n ¡ Information theoretic measures, KL-divergence, etc. Transduction is not global! Most substantial gain with limited labeled data Current work in progress: ¡ TSVMs n n ¡ Improve performance on smaller, unbalanced data Select an improved smaller dimensional feature space representation Ensemble classifiers, Bayesian model averaging, Mixture of experts 6
- Slides: 6