Seeded Bayesian Networks Constructing Genetic Networks From Microarray
Seeded Bayesian Networks: Constructing Genetic Networks From Microarray Data By: Amira Djebbari and John Quackenbush BMC Systems Biology 2008, 2: 57 Presented by: Garron Wright April 20, 2009 CSCE 582

Background • Bioinformatics • Gene Network Modeling Techniques • Microarrays • Seeded Bayesian Networks/a priori knowledge biasing
Bioinformatics • Confluence of biology, computer science, and information technology • Ultimate goal: enable the discovery of new biological insights as well as creating global perspective that elucidates unifying principles of biology • Genetic information is then used to create a comprehensive picture of cellular function in normal state to then compare to diseased and other altered states • Interpretation of nucleotide sequence data, amino acid sequence data, protein domains, and protein structures Source: http: //www. ncbi. nlm. nih. gov
Microarrays Source: http: //www. bio. davidson. edu/Courses/genomics/chip. html
Microarrays
Gene Network Modeling Techniques • Weighted matrices • Boolean Networks • Differential Equations • Bayesian Networks

Cellular Complexity: An NP-Complete Problem • All potential network topologies must be assessed • Possible Solution: Heuristic Search Algorithms • Example: Greedy Hill Climbing • Problem: Often find local maxima but not global
Domain-Specific Knowledge in Constructing Bayesian Neworks • Leads to near optimization in exploring the state space (relative gene expression state in this instance) • Network seed biases the search for the best topology, however it does not limit novel gene interactions from being identified • How do we seed/set a priori knowledge? • Pathway/interaction databases • Networks deduced from published literature in Pub. Med • High-throughput interaction screens (PPI)

Procedure • Deducing Prior Knowledge From Published Literature • Co-occurrence method • Limited papers/literature to just 2 genes thus exhibits scale-free behavior • Deducing Prior Network Structure From High-Throughput Screens • Interactome data • PPI • Represent unbiased screens for interactions and have shown new unreported interactions • Modified Depth-First Search to imply directionality to the network structure and then bootstrapping 100 times for the following cases: no priors, literature derived priors, PPI derived priors, and a combination of the two • Select features with bootstrap confidence of 0. 7 or greater
Procedure (cont. ) • Bayesian Network analysis of gene expression on a leukemia study • Comparison of Acute Lymphoblastic Lymphoma (ALL) to Acute Myeloid Leukemia (AML) • Top 40 genes that distinguish between the two cancers • Microarray does not probe entire genome, only a subset • Bayesian analysis of gene expression on a second leukemia study • Nearly the entire genome probed • Evaluated network reconstruction on the KEGG cell cycle pathway • Created a Receiver-Operator (ROC) Curve (True positive rate vs. False Positive rate)
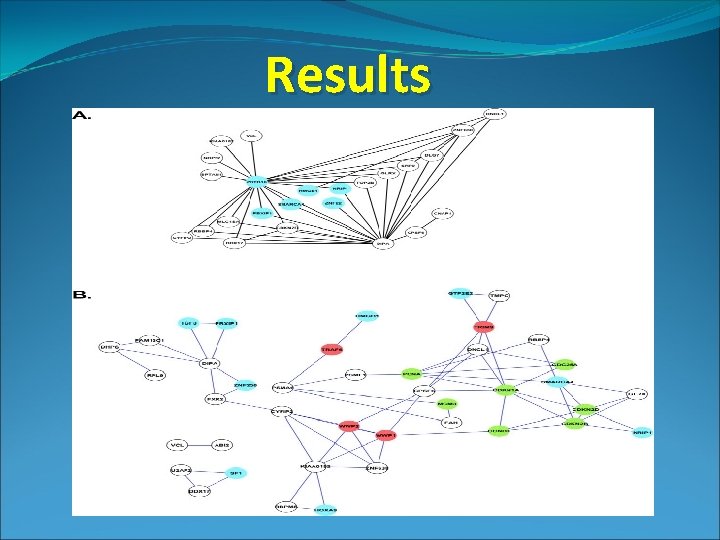
Results




Discussion • Advent of microarray and other high throughput technology led to the expectation that the pathways/networks that would link genotype to phenotype would be discovered • Using BN analysis as described here can recover some of that promise • The two gene expression data sets are typical of microarray studies • Domain specific knowledge of prior network seeds improves the ability of BN learning the interactions between genes • These interactions can be used to reconstruct predictive networks at any confidence level • Automated way to derive network graphs from a gene list, refining the graph with the expression data, and learning conditional probabilities that can be used to predict gene response to various insults • Wide range of uses: mechanistic studies to drug target
- Slides: 17