Section II Protein Structure Linear sequence Spatial structure






- Slides: 54

Section II Protein Structure Linear sequence ? Spatial structure

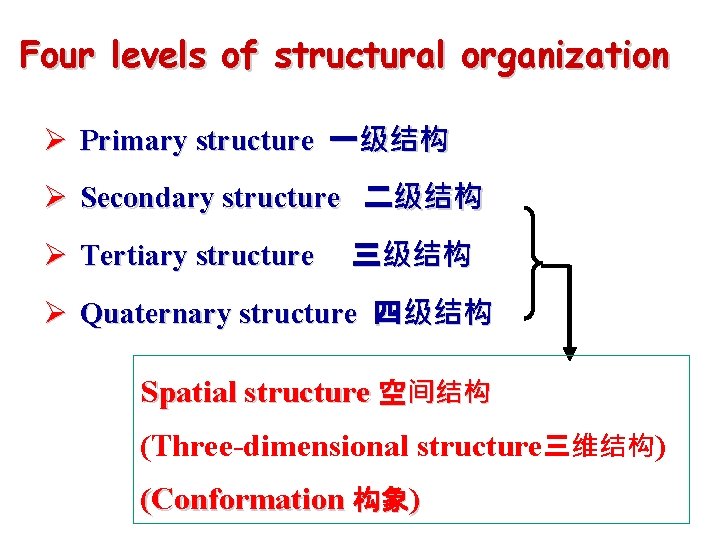
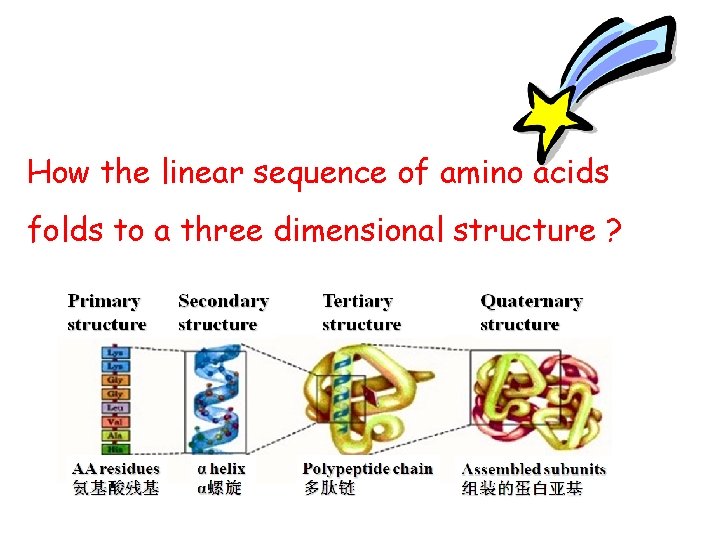
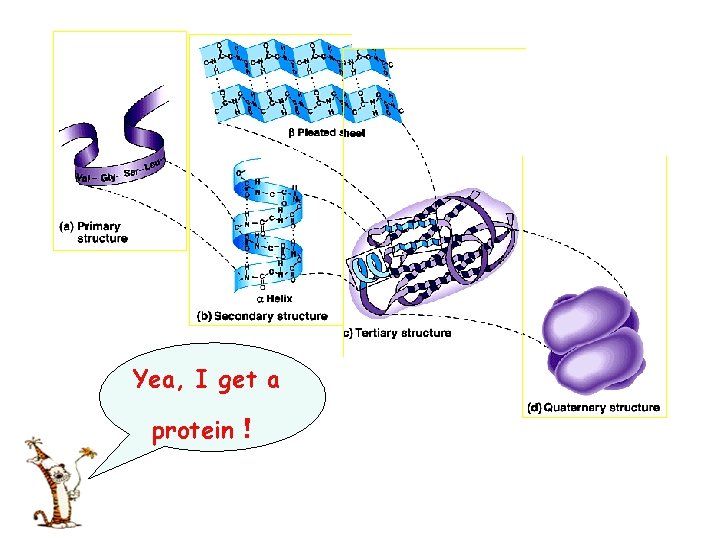
Four levels of structural organization Ø Primary structure 一级结构 Ø Secondary structure 二级结构 Ø Tertiary structure 三级结构 Ø Quaternary structure 四级结构 Spatial structure 空间结构 (Three-dimensional structure三维结构) (Conformation 构象)

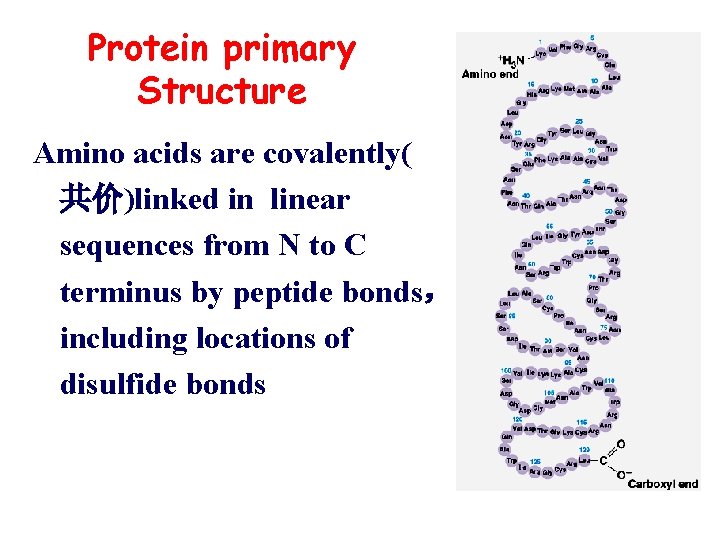
Protein primary Structure Amino acids are covalently( 共价)linked in linear sequences from N to C terminus by peptide bonds, including locations of disulfide bonds

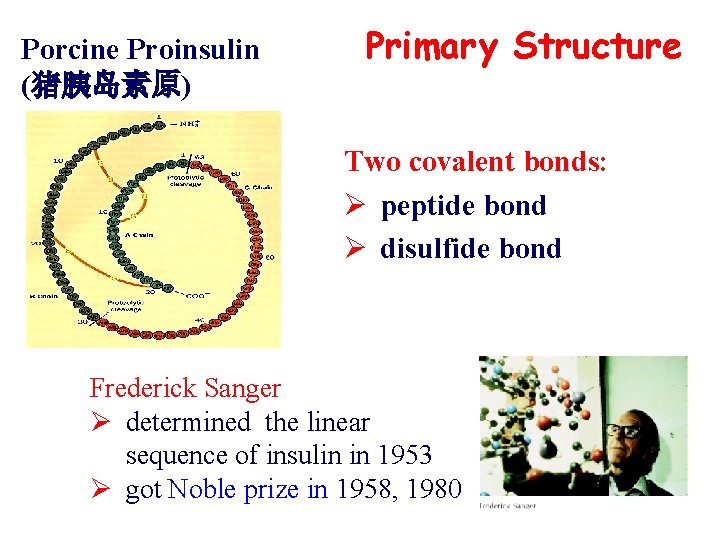
Porcine Proinsulin (猪胰岛素原) Primary Structure Two covalent bonds: Ø peptide bond Ø disulfide bond Frederick Sanger Ø determined the linear sequence of insulin in 1953 Ø got Noble prize in 1958, 1980

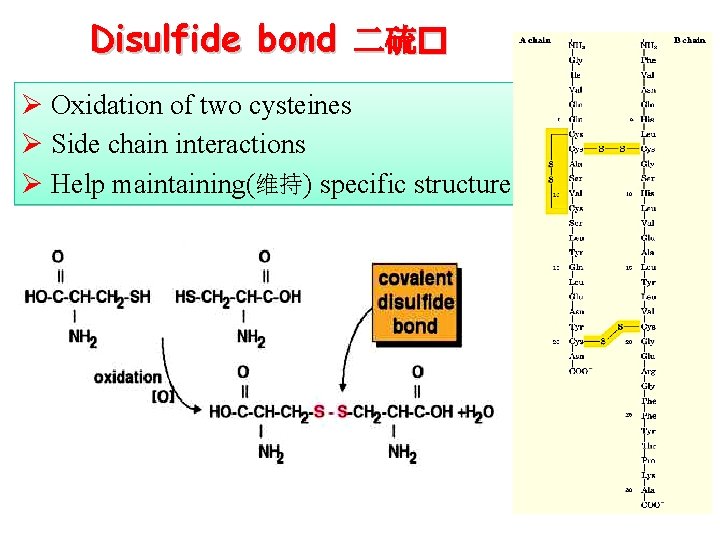
Disulfide bond 二硫� Ø Oxidation of two cysteines Ø Side chain interactions Ø Help maintaining(维持) specific structure

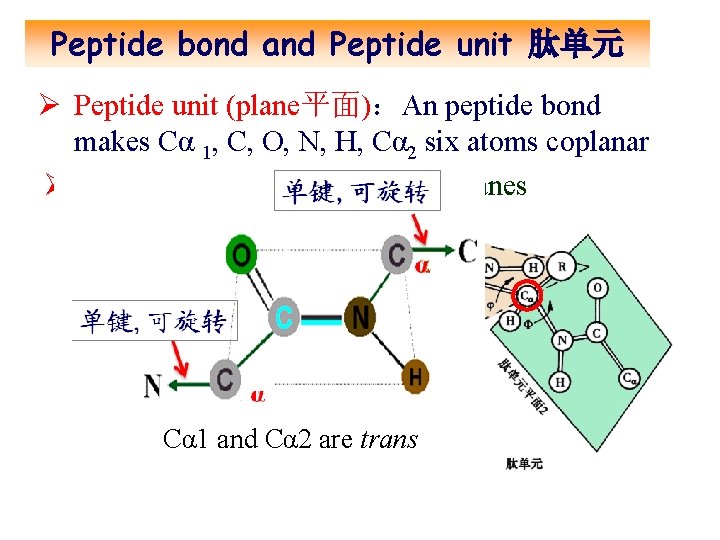
Peptide bond and Peptide unit 肽单元 Ø Peptide unit (plane平面):An peptide bond makes Cα 1, C, O, N, H, Cα 2 six atoms coplanar Ø Cα is the joining point for two planes Cα 1 and Cα 2 are trans

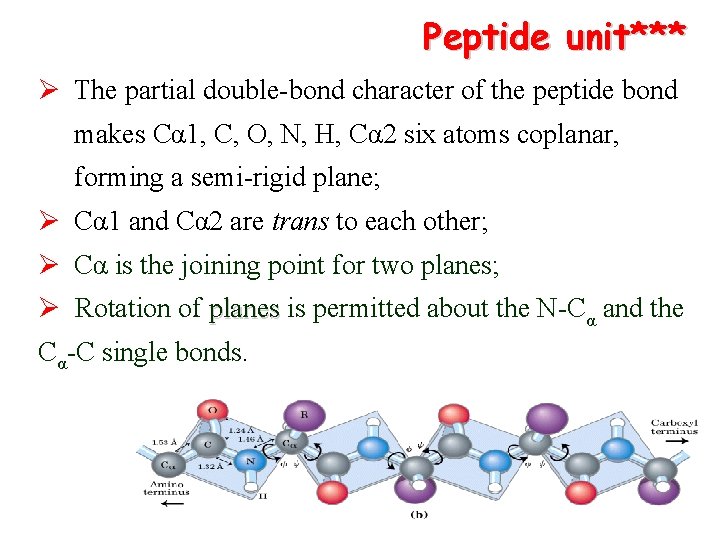
Peptide unit*** Ø The partial double-bond character of the peptide bond makes Cα 1, C, O, N, H, Cα 2 six atoms coplanar, forming a semi-rigid plane; Ø Cα 1 and Cα 2 are trans to each other; Ø Cα is the joining point for two planes; Ø Rotation of planes is permitted about the N-Cα and the Cα-C single bonds.

Summary: Primary structure Ø The AA sequence from N end to C end in the polypeptide chain(s) of a protein Ø Number, sort and order (AA的数目, 种类和顺序) Ø Forces: covalent bonds—peptide bonds and disulfide bonds (if present) Ø The AA sequence of a protein is determined by its gene 氨基酸的序列由其基因决定

How the linear sequence of amino acids folds to a three dimensional structure ?

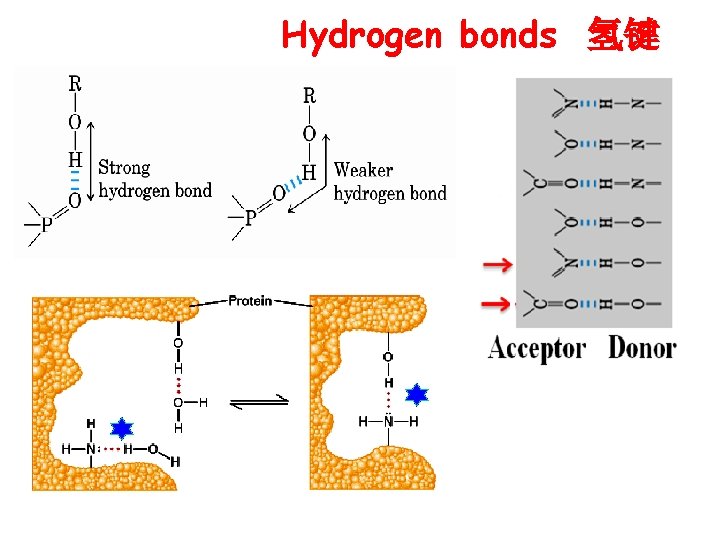
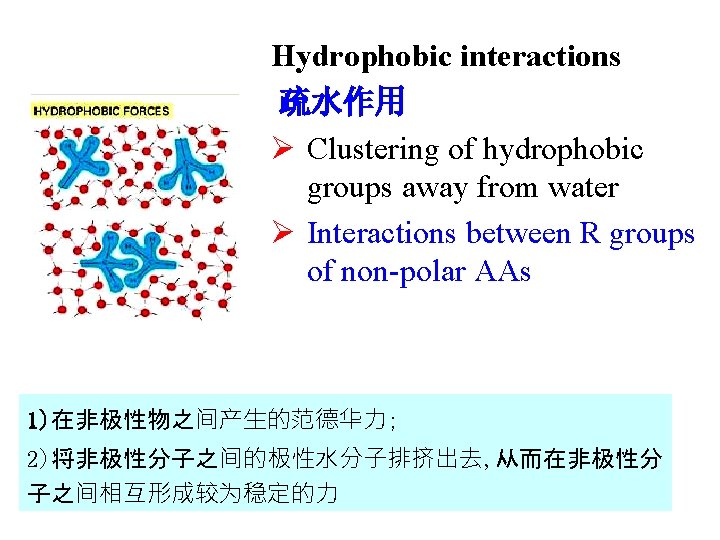
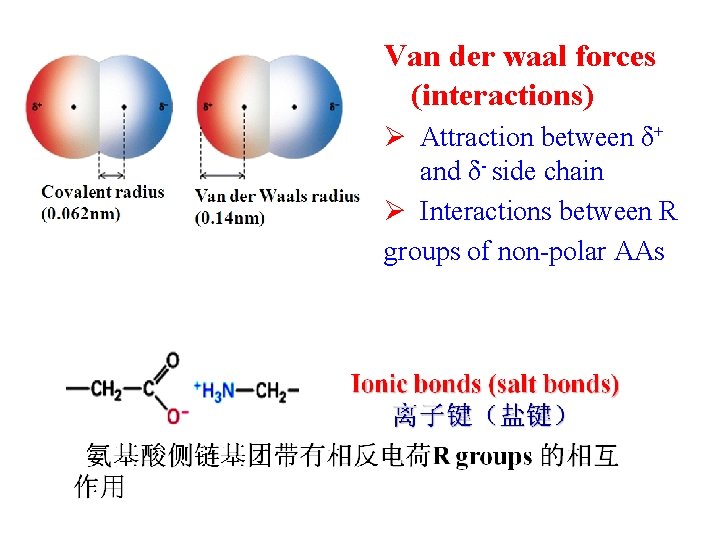
Non-covalent interactions (非共价作用力) in spatial structure Ø hydrogen bonds (氢键) Ø hydrophobic interactions (疏水作用) Ø ionic bonds (离子键) Ø Van der waals forces (范德华力)

Hydrogen bonds 氢键

Hydrophobic interactions 疏水作用 Ø Clustering of hydrophobic groups away from water Ø Interactions between R groups of non-polar AAs 1)在非极性物之间产生的范德华力; 2)将非极性分子之间的极性水分子排挤出去, 从而在非极性分 子之间相互形成较为稳定的力

Van der waal forces (interactions) Ø Attraction between δ+ and δ- side chain Ø Interactions between R groups of non-polar AAs

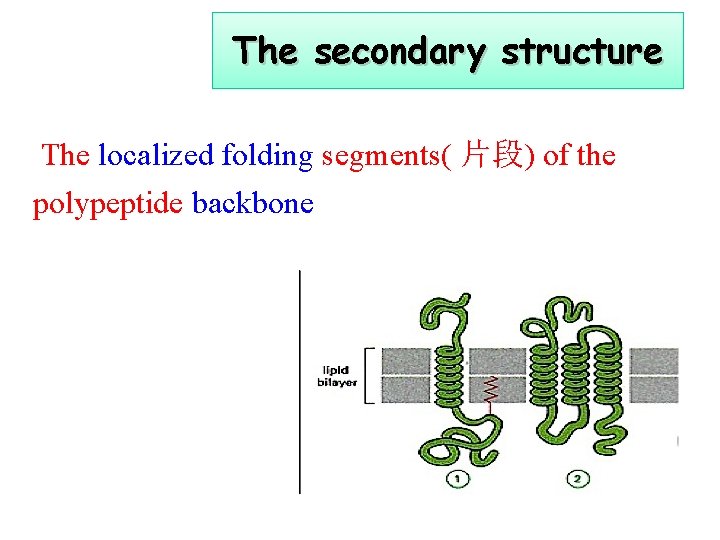
The secondary structure The localized folding segments( 片段) of the polypeptide backbone

The secondary structure Ø Common types of the secondary structure***: α-helix (α螺旋) β-pleated sheet (β折叠) Random coil (无规卷曲) β-turn(bend)(β转角) Ø Forces: hydrogen bonds (mainly)

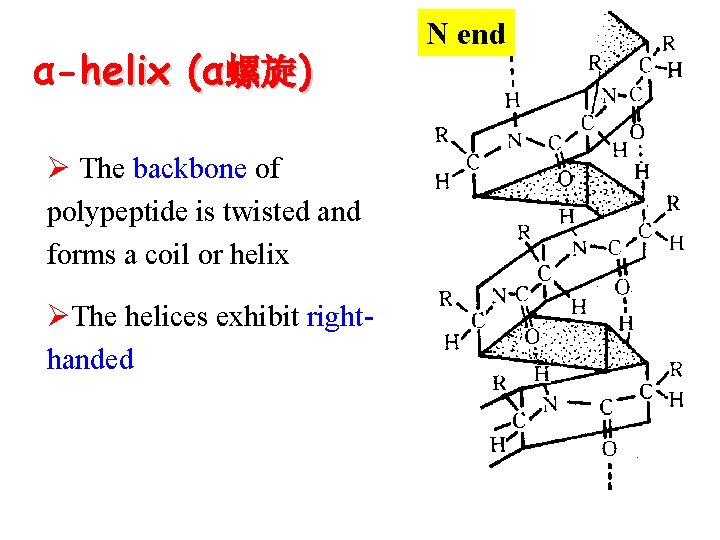
α-helix (α螺旋) Ø The backbone of polypeptide is twisted and forms a coil or helix ØThe helices exhibit righthanded N end

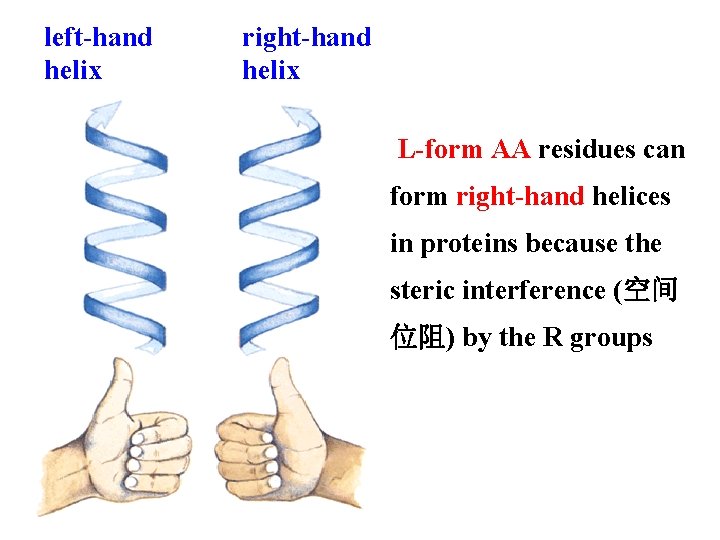
left-hand helix right-hand helix L-form AA residues can form right-hand helices in proteins because the steric interference (空间 位阻) by the R groups

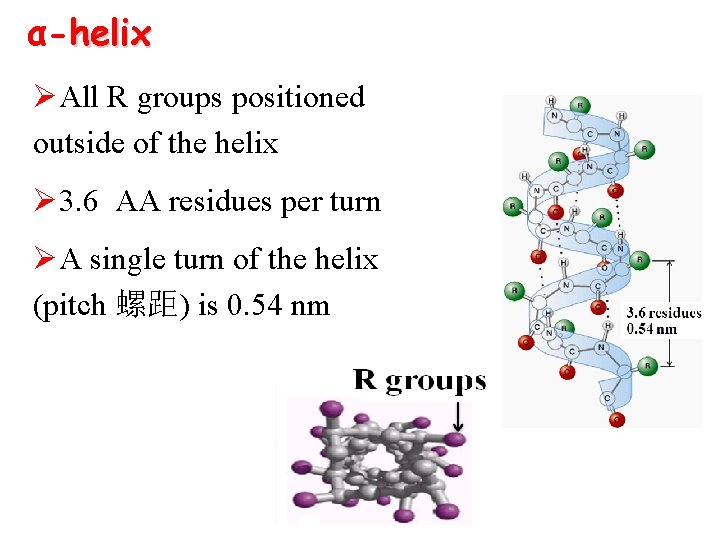
α-helix ØAll R groups positioned outside of the helix Ø 3. 6 AA residues per turn ØA single turn of the helix (pitch 螺距) is 0. 54 nm

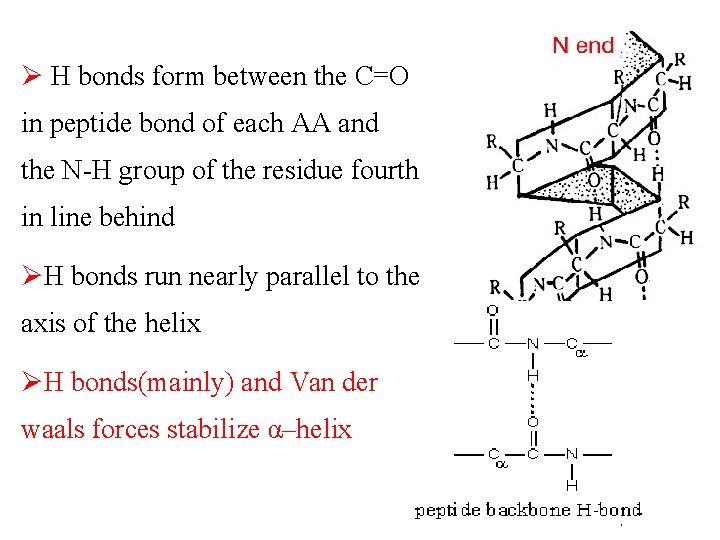
Ø H bonds form between the C=O in peptide bond of each AA and the N-H group of the residue fourth in line behind ØH bonds run nearly parallel to the axis of the helix ØH bonds(mainly) and Van der waals forces stabilize α–helix

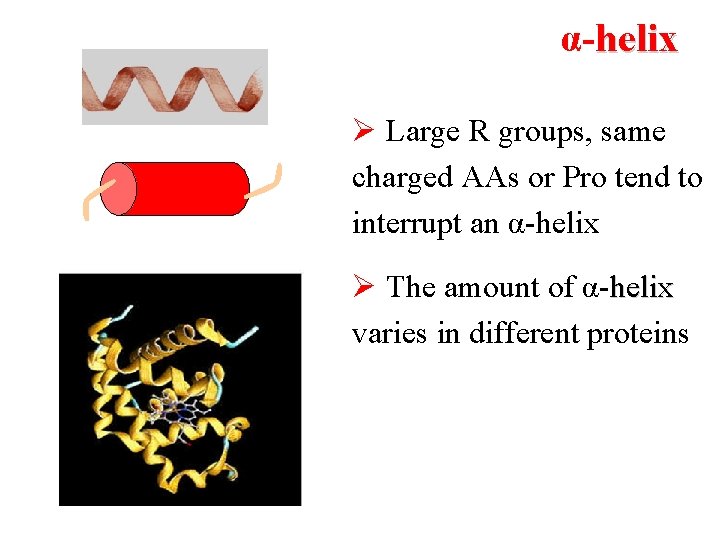
α-helix Ø Large R groups, same charged AAs or Pro tend to interrupt an α-helix Ø The amount of α-helix varies in different proteins

Summary: α-helix Ø Backbone:forms a right-handed helix Ø Side chain:all R groups positioned outside of the helix Ø One turn:contains 3. 6 AA with 0. 54 nm high Ø Force: hydrogen bond that is parallel to helix axis is the main force to stabilize the helix Ø All peptide bonds in a helix are involved in formation of H bonds

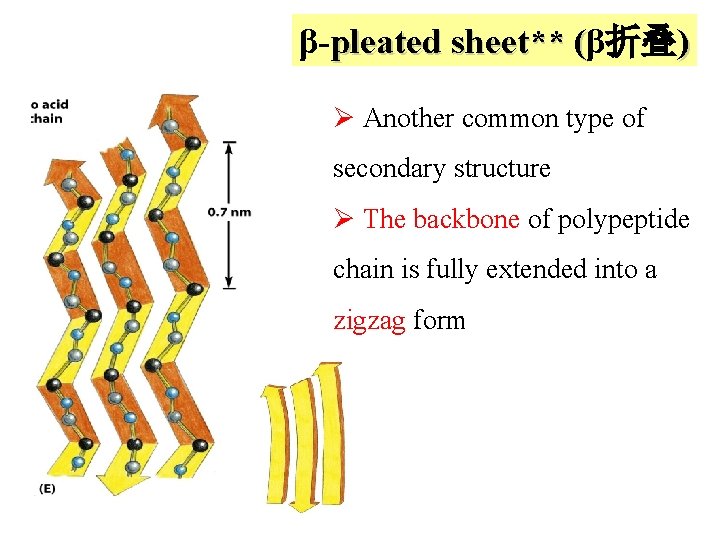
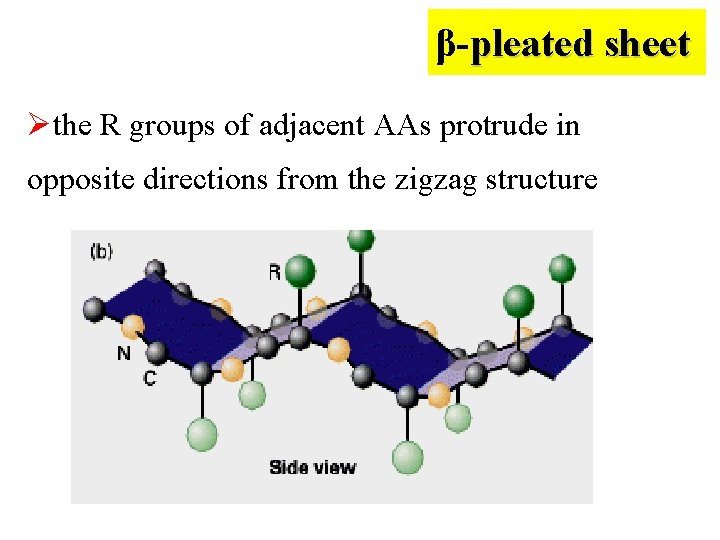
β-pleated sheet** (β折叠) Ø Another common type of secondary structure Ø The backbone of polypeptide chain is fully extended into a zigzag form

β-pleated sheet Øthe R groups of adjacent AAs protrude in opposite directions from the zigzag structure

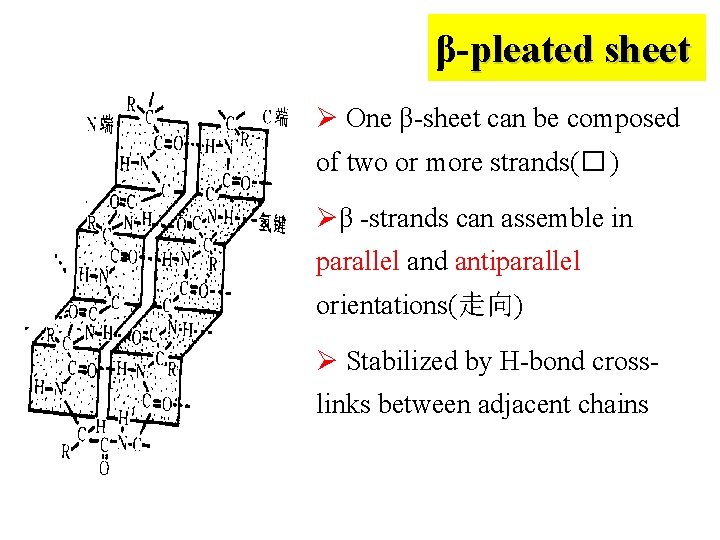
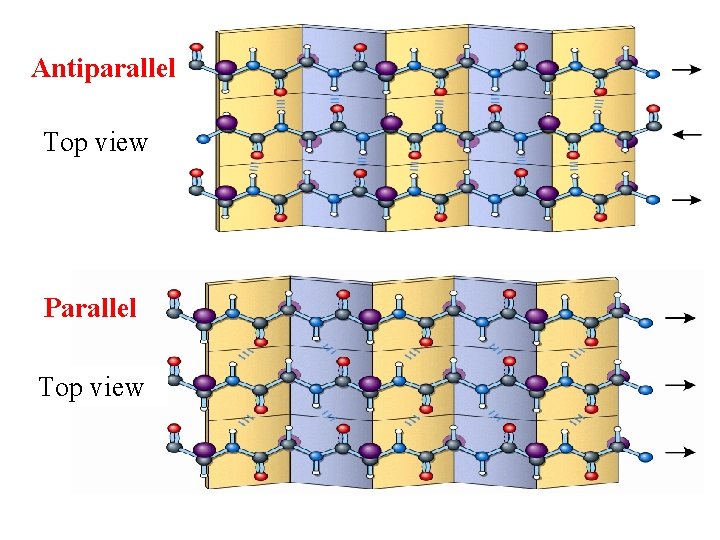
β-pleated sheet Ø One β-sheet can be composed of two or more strands(� ) Øβ -strands can assemble in parallel and antiparallel orientations(走向) Ø Stabilized by H-bond crosslinks between adjacent chains

Antiparallel Top view Parallel Top view

β-pleated sheet

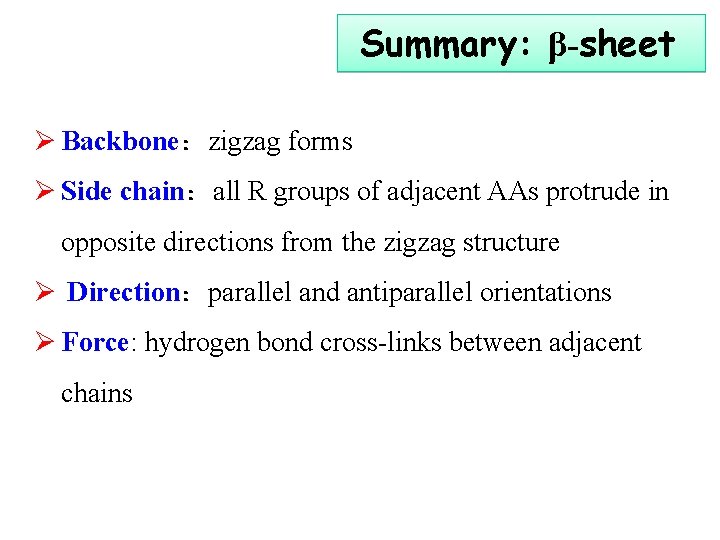
Summary: β-sheet Ø Backbone:zigzag forms Ø Side chain:all R groups of adjacent AAs protrude in opposite directions from the zigzag structure Ø Direction:parallel and antiparallel orientations Ø Force: hydrogen bond cross-links between adjacent chains

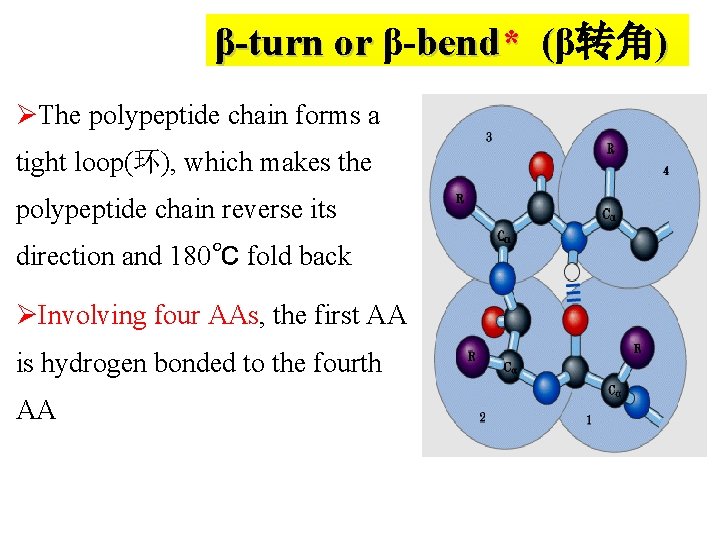
β-turn or β-bend* (β转角 ) ( ØThe polypeptide chain forms a tight loop(环), which makes the polypeptide chain reverse its direction and 180℃ fold back ØInvolving four AAs, the first AA is hydrogen bonded to the fourth AA

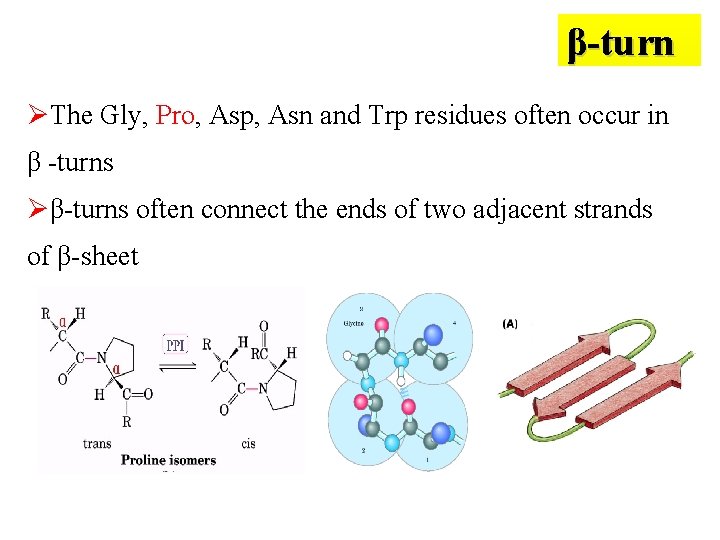
β-turn ØThe Gly, Pro, Asp, Asn and Trp residues often occur in β -turns Øβ-turns often connect the ends of two adjacent strands of β-sheet

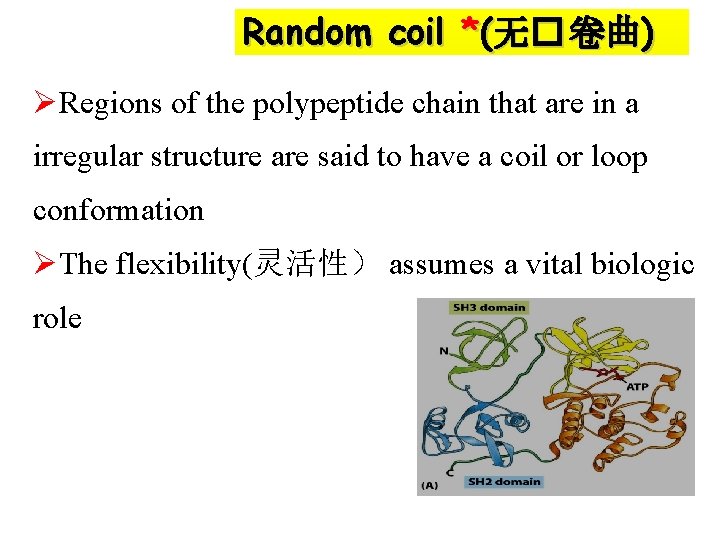
Random coil *(无� 卷曲) ØRegions of the polypeptide chain that are in a irregular structure are said to have a coil or loop conformation ØThe flexibility(灵活性) assumes a vital biologic role

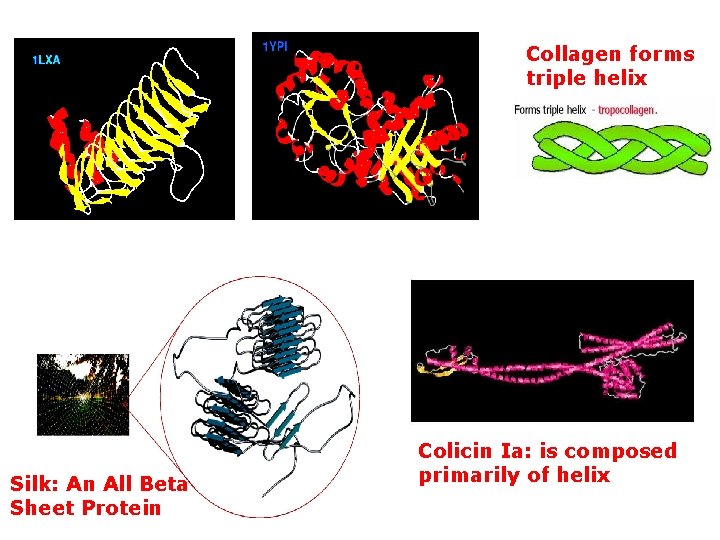
Collagen forms triple helix Silk: An All Beta Sheet Protein Colicin Ia: is composed primarily of helix

Summary: the secondary structure Ø Reasons for the formation -- semi-rigid peptide bond plane -- R groups: charge, size, steric interference Ø A result of H-bonding between peptide bonds of AAs within the protein Ø nearly all proteins contain either α -helices, β -sheets or both

Summary: the secondary structure Ø Addition of new properties to a protein like strength, flexibility, ∙∙∙ -- α-helices: tough, insoluble protective structures of varying hardness and flexibility -- β-sheet: soft, flexible filaments (细丝)

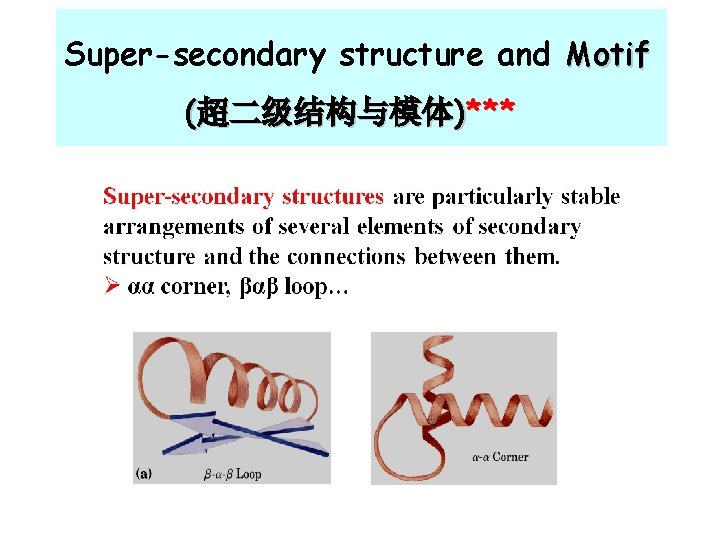
Super-secondary structure and Motif (超二级结构与模体)***

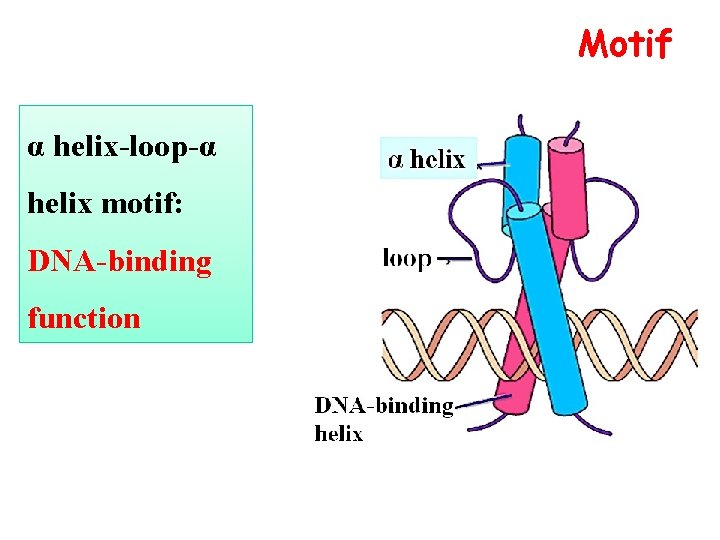
Motif α helix-loop-α helix motif: DNA-binding function

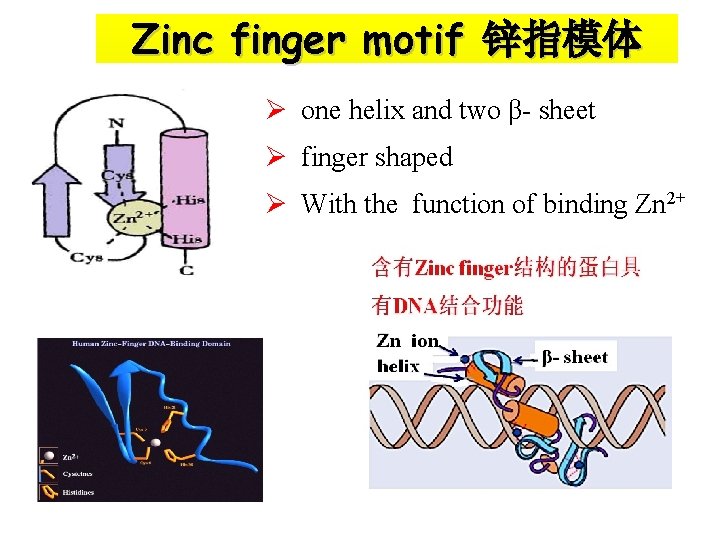
Zinc finger motif 锌指模体 Ø one helix and two β- sheet Ø finger shaped Ø With the function of binding Zn 2+

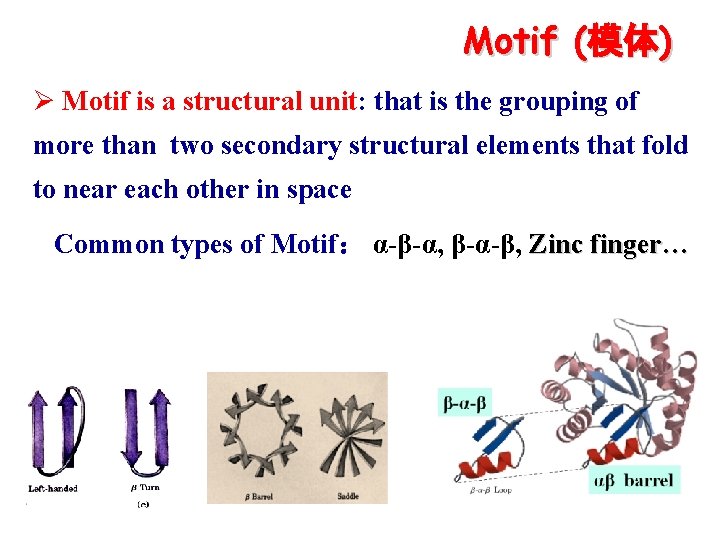
Motif (模体) Ø Motif is a structural unit: that is the grouping of more than two secondary structural elements that fold to near each other in space Common types of Motif: α-β-α, β-α-β, Zinc finger…

Motif Ø Motif is a functional unit: it is essential for a motif with a special function e. g. Zinc finger motif exerts DNA-binding activity ØSome motifs consist of only a few conserved(保守 的) functionally important AAs rather than supersecondary structures e. g. RGD (Arg-Gly-Asp ) motif 纤连蛋白与其受体结合区域

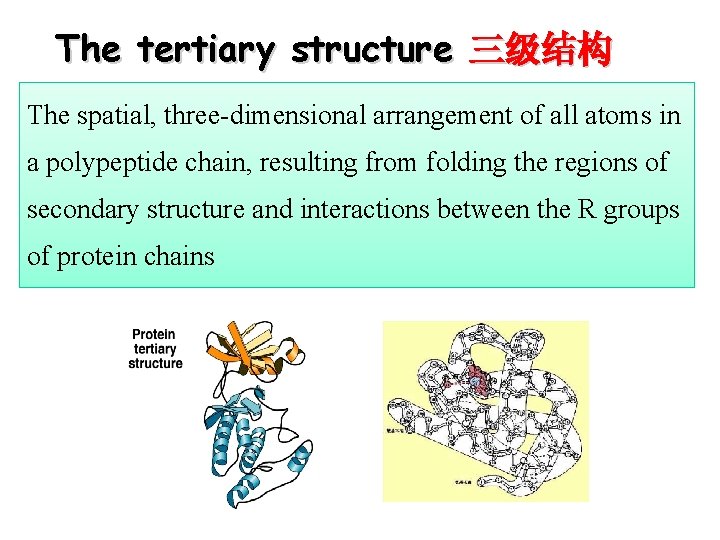
The tertiary structure 三级结构 The spatial, three-dimensional arrangement of all atoms in a polypeptide chain, resulting from folding the regions of secondary structure and interactions between the R groups of protein chains

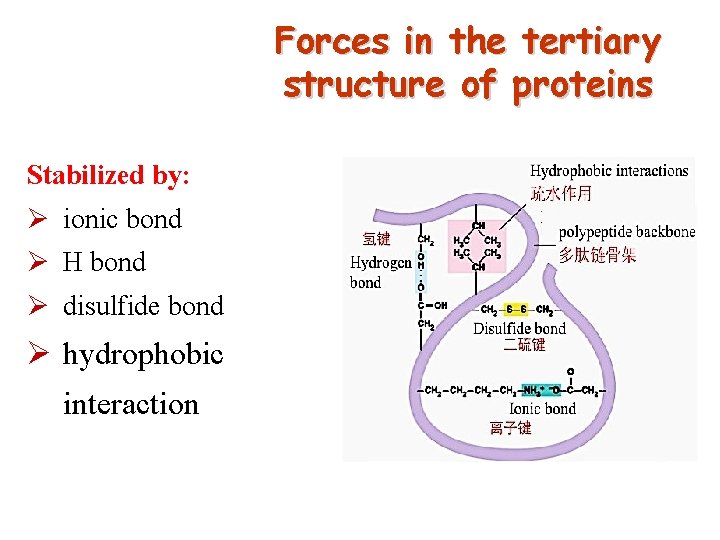
Forces in the tertiary structure of proteins Stabilized by: Ø ionic bond Ø H bond Ø disulfide bond Ø hydrophobic interaction

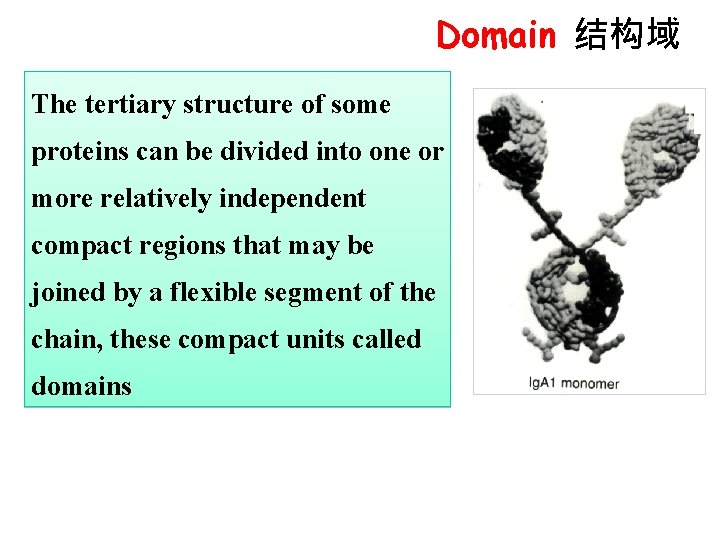
Domain 结构域 The tertiary structure of some proteins can be divided into one or more relatively independent compact regions that may be joined by a flexible segment of the chain, these compact units called domains

Domain ØA distinct structural unit within a large polypeptide, may compose several motifs --domain > motif, composed of 40 -400 AAs ØA functional unit, the individual domains have separate functions Ø One protein may contain several domains

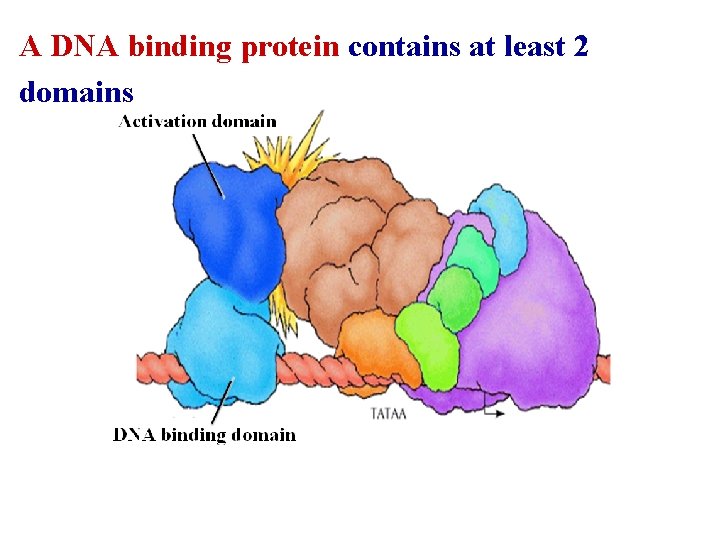
A DNA binding protein contains at least 2 domains

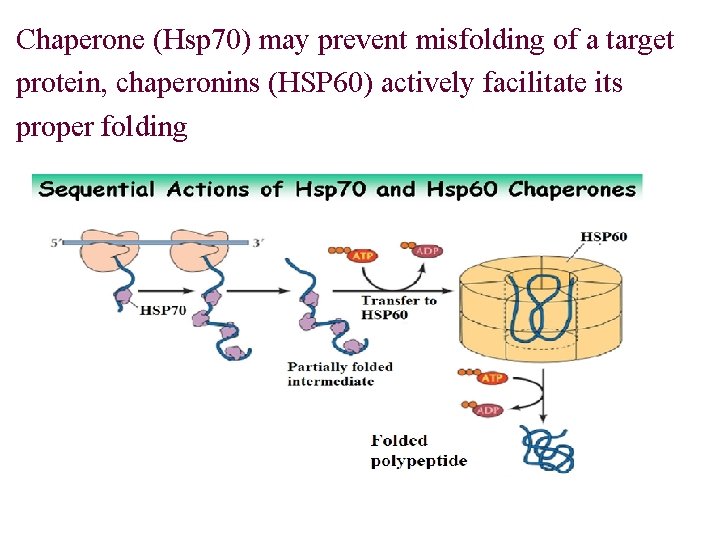
Polypeptide Chain Folding 肽链折叠 Ø Proteins will be at the lowest free energy state when they fold to a correct or “native” state Ø The information for folding resides within its AA sequence 折叠信息储存在蛋白质的一级结构中 Ø Folding is self-assemble(组装), or assisted by some of specific proteins: chaperones(分子伴侣), chaperonins(伴侣素) and others

Chaperone (Hsp 70) may prevent misfolding of a target protein, chaperonins (HSP 60) actively facilitate its proper folding

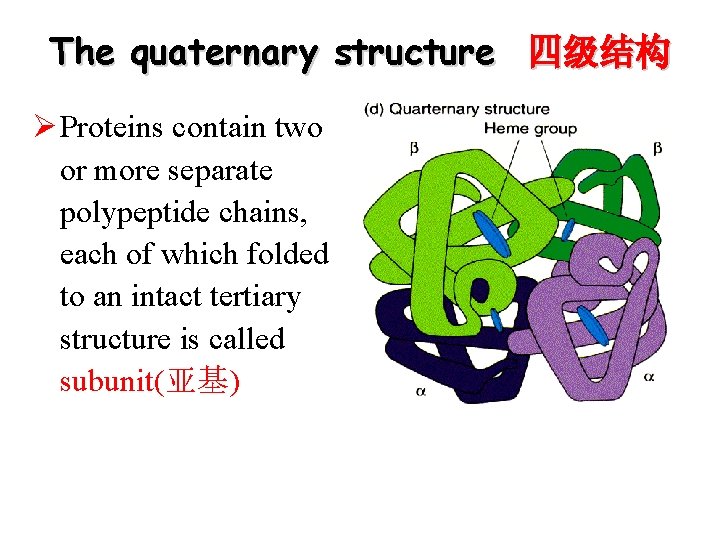
The quaternary structure 四级结构 Ø Proteins contain two or more separate polypeptide chains, each of which folded to an intact tertiary structure is called subunit(亚基)

The quaternary structure Ø Association of two or more subunits to form a functional protein with quaternary structure Ø Subunits can be identical(相同的) or not homomultimeric 同多聚体: one kind of subunits heteromultimeric 异多聚体: several different kinds of subunits

Forces involved in quaternary structure Ø hydrogen bonds (mainly) Ø ionic bonds (mainly) Ø Van der waals Ø hydrophobic forces Ø Disulfide bonds(if present) 亚基之间的结合力主要是氢键和离子键

Yea, I get a protein!

Factors determine protein spatial structures ØThe primary structure :basis of high spatial structure of a protein ØInteractions ØThe with solvent molecules p. H and ionic composition of the solvent

Classification of proteins (自学) Based on compositions: Ø Simple proteins 简单蛋白: Contain AAs only serum albumin(清蛋白), keratin(角蛋白) Ø Conjugated proteins 结合蛋白: ---metalloproteins: protein+metal ions ---glycoproteins: protein + carbohydrate group ---lipoproteins: protein + lipid molecules

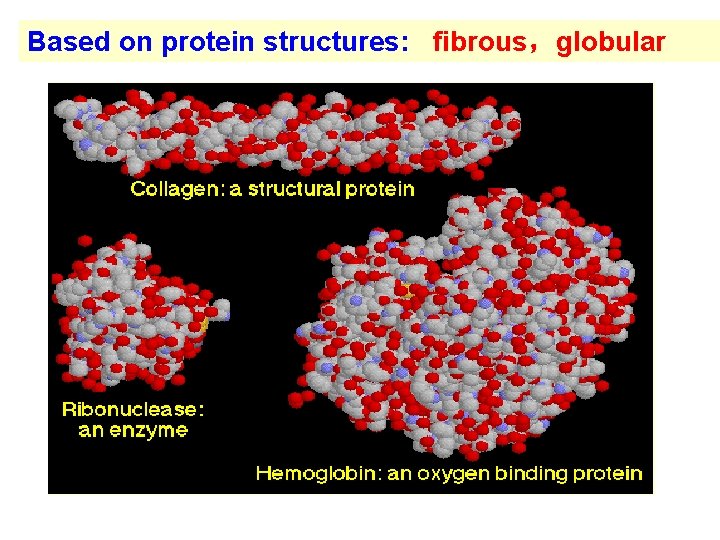
Based on protein structures: fibrous,globular

Protein functions ØEnzymatic proteins selective acceleration of chemical reactions-- enzymes ØRegulatory proteins control metabolism and gene expression-transcription factors ØTransport proteins transport of other substances-- blood gas transport ØStorage proteins: storage of AAs-- egg albumin

Protein functions ØContractile proteins Movement-- muscle, actin(肌动蛋白), cilia (睫毛) ØSupport Fibrous framework in animal connective tissues-- collagen, keratin, silk ØDefensive proteins Protection against disease-- antibodies