Scientific Benchmarks for Structure Prediction Codes Jack Snoeyink
Scientific Benchmarks for Structure Prediction Codes Jack Snoeyink & Matt O’Meara Dept. Computer Science UNC Chapel Hill

With thanks to: Collaborators � Brian Kuhlman, UNC Biochem � Many other members of the Rosetta. Commons � Richardson lab, Duke Biochem Funding � NIH � NSF
Key Points… � Scientific Models, esp. for Structural Molecular Biology � � � Focus on statistical/computational models with � � Models are the lens through which we view data Models are predominantly geometric Computational models are complex Models evolve, so testing becomes crucial a sample source, observable local features, chosen functional form, fit parameters, & visualization/testing methods Capture assumptions and date used to build models to: � � � Visualize for making design decisions while building Fit parameters to ensure best performance Record as scientific benchmarks Case Study: Rosetta protein structure prediction software [B]

Science views nature thru models

Scientists view nature thru models

People view the world thru models
Geometric molecular models
Model complexity � Physical and Conceptual models � � Kept simple to aid understanding Statistical and Computational models Evolve by combining simple models � Even when complex can still be effective at Validation (Molprobity) or Prediction (Rosetta) �
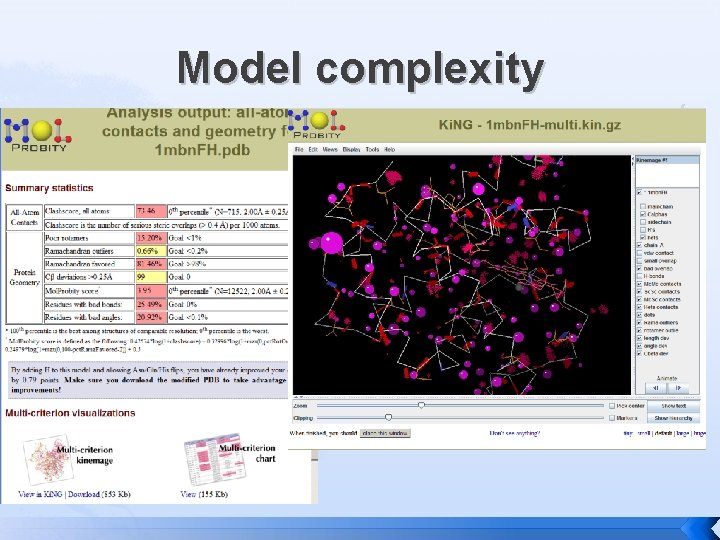
Model complexity
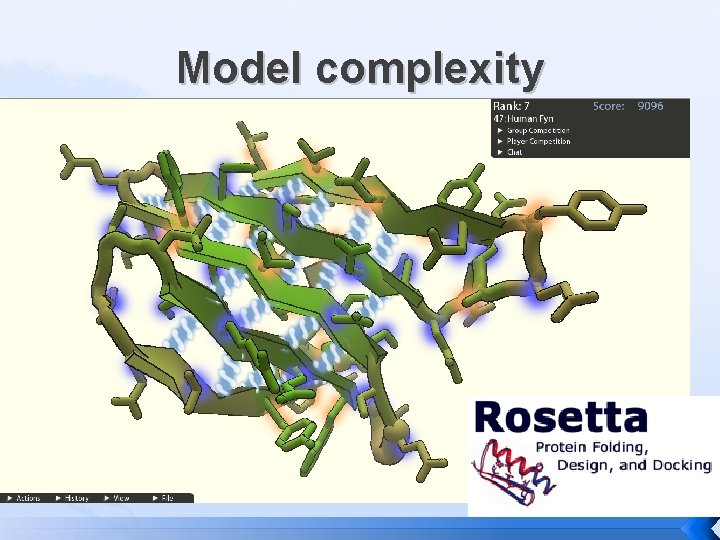
Model complexity
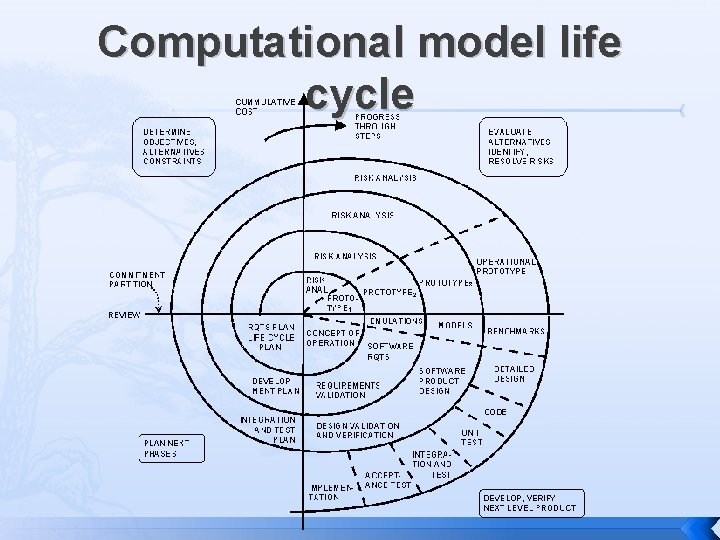
Computational model life cycle
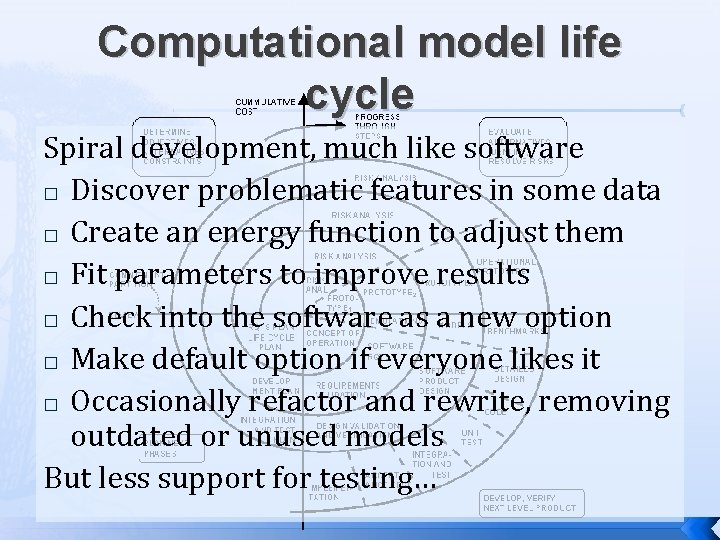
Computational model life cycle Spiral development, much like software � Discover problematic features in some data � Create an energy function to adjust them � Fit parameters to improve results � Check into the software as a new option � Make default option if everyone likes it � Occasionally refactor and rewrite, removing outdated or unused models But less support for testing…
Computational model testing Our goal: Capture data and assumptions from model building for use in model visualization and testing.
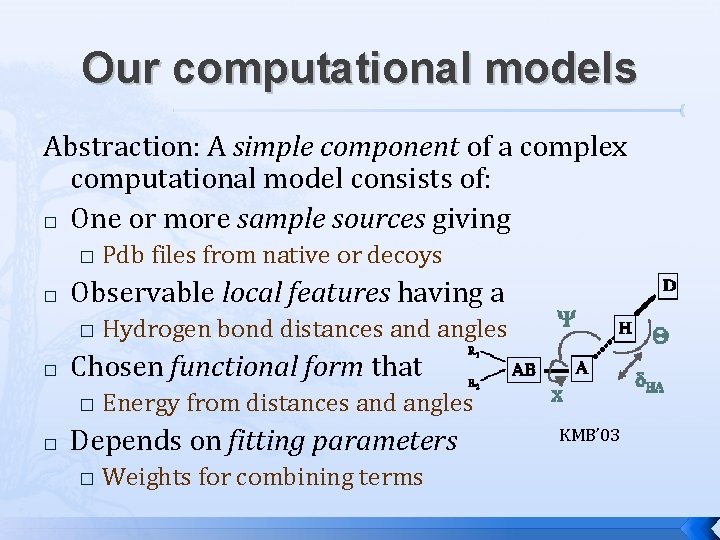
Our computational models Abstraction: A simple component of a complex computational model consists of: � One or more sample sources giving � � Observable local features having a � � Hydrogen bond distances and angles Chosen functional form that � � Pdb files from native or decoys Energy from distances and angles Depends on fitting parameters � Weights for combining terms KMB’ 03
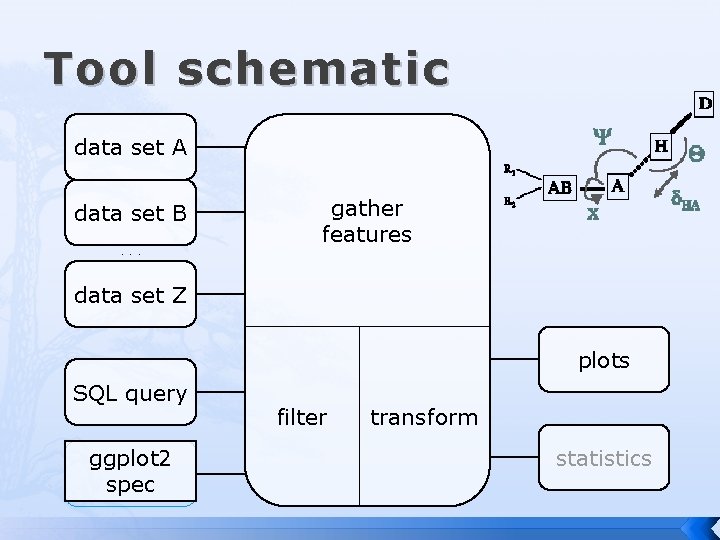
Tool schematic data set A data set B. . . gather features data set Z plots SQL query ggplot 2 spec filter transform statistics
Visualization Implemented tools � Compare distributions from sample sources � Tufte’s small multiples via ggplot � Kernel density estimation � Normalization Opportunities for � Statistical analysis � Dimension reduction …
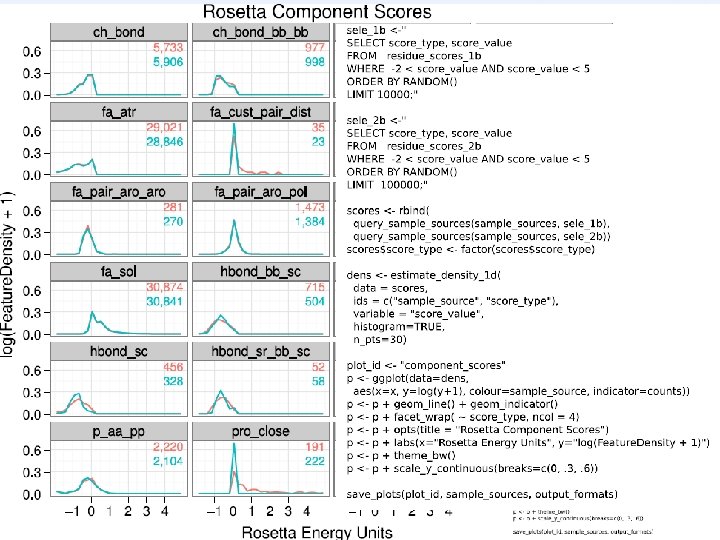
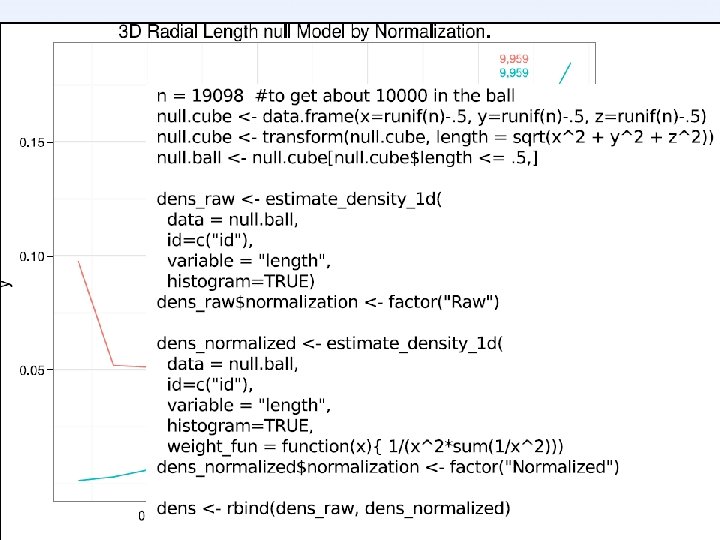
![Normalization [KMB’ 03] Histogram of Hbond A-H distances in natives Normalization [KMB’ 03] Histogram of Hbond A-H distances in natives](http://slidetodoc.com/presentation_image_h2/d4c400ad0e386b40922985ab3de5ddda/image-18.jpg)
Normalization [KMB’ 03] Histogram of Hbond A-H distances in natives
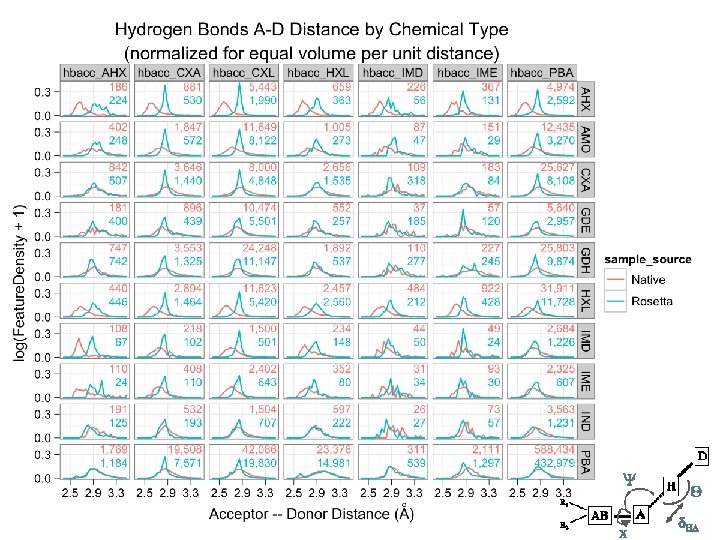
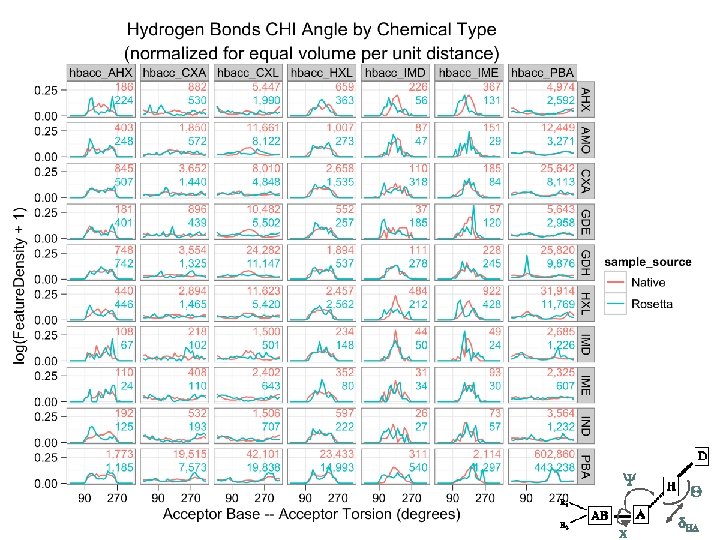
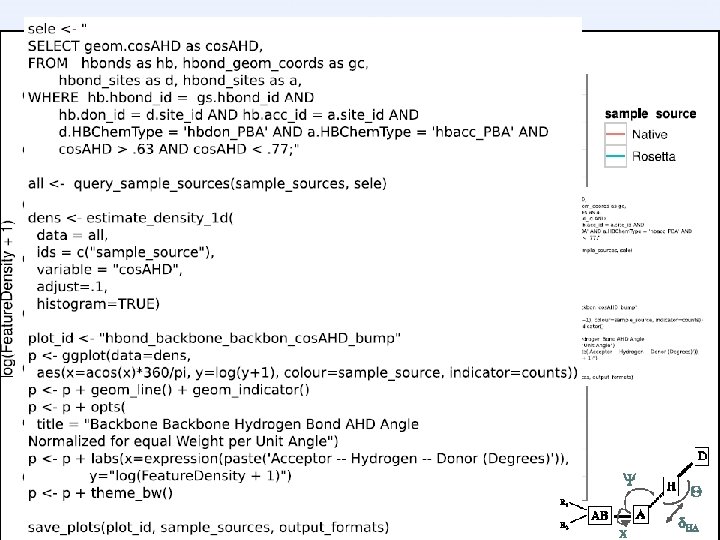

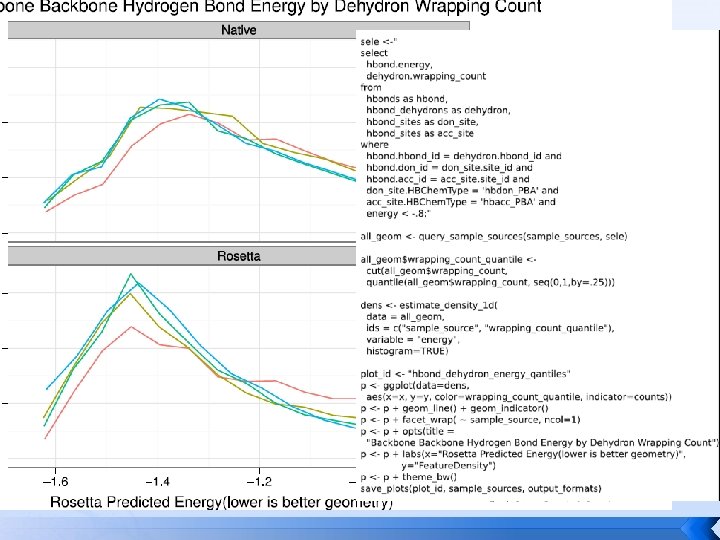
Tool uses… Scientific unit tests native, HEAD, ^HEAD run on continuously testing server Knowledge-base score term creation native, release, experimental turn exploration into living benchmarks Test design hypotheses native, protocol, designs how strange is the this geometry?
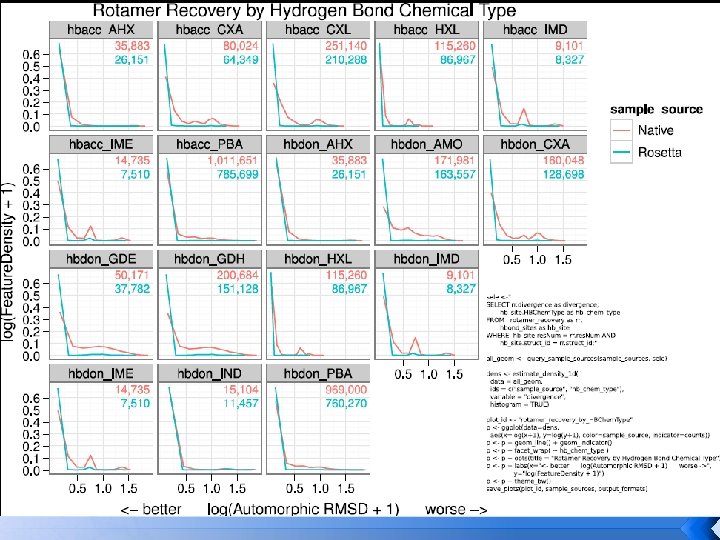
Rotamer recovery
Key Points… � Scientific Models, esp. for Structural Molecular Biology � � � Focus on statistical/computational models with � � Models are the lens through which we view data Models are predominantly geometric Computational models are complex Models evolve, so testing becomes crucial a sample source, observable local features, chosen functional form, fit parameters, & visualization/testing methods Capture assumptions and date used to build models to: � � � Visualize for making design decisions while building Fit parameters to ensure best performance Record as scientific benchmarks Case Study: Rosetta protein structure prediction software [B]
- Slides: 27