Schedule Lecture 4 Recap Lecture 5 pAcceptor Ligands






- Slides: 18


Schedule • Lecture 4: Re-cap • Lecture 5: p-Acceptor Ligands and Biology CO and O 2 complexes • Lecture 6: N 2 and NO complexes, M-M bonds More on p-acceptor ligands, introduction to metal-metal bonding Slide 2/18

Summary of the Last Lecture Metal-carbonyl complexes • Bonding is due to synergic OC M s-donation and M CO p-back donation • Reduction in vco stretching frequency is related to the extent of back-bonding • Number of v. CO in IR and Raman can be used to work out structure O 2 complexes • Haemoglobin and myoglobin bind weakly to O 2 allowing transport and storage of highly reactive molecule Today’s lecture • N 2 complexes and Metal-Metal bonding Slide 3/18

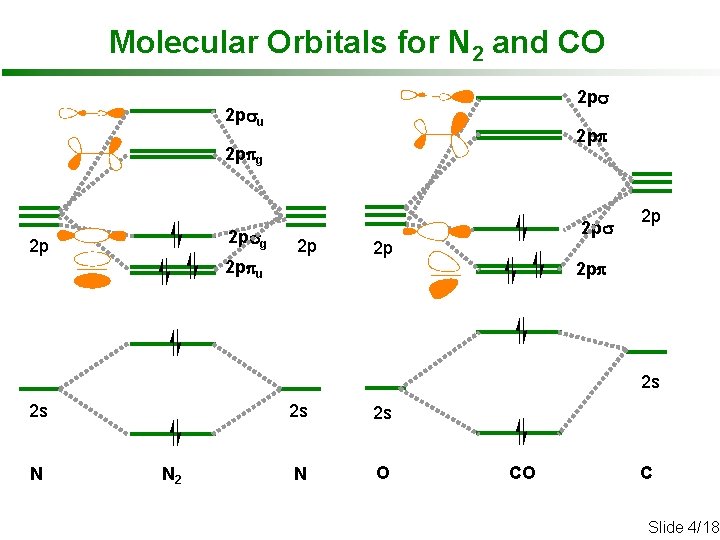
Molecular Orbitals for N 2 and CO 2 psu 2 ppg 2 psg 2 p 2 p 2 ppu 2 ps 2 p 2 p 2 pp 2 s 2 s N N 2 2 s 2 s N O CO C Slide 4/18

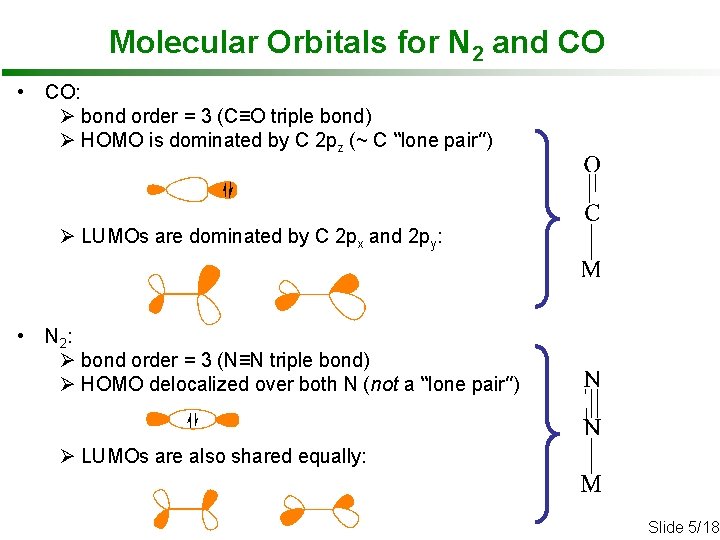
Molecular Orbitals for N 2 and CO • CO: Ø bond order = 3 (C≡O triple bond) Ø HOMO is dominated by C 2 pz (~ C “lone pair”) Ø LUMOs are dominated by C 2 px and 2 py: • N 2 : Ø bond order = 3 (N≡N triple bond) Ø HOMO delocalized over both N (not a “lone pair”) Ø LUMOs are also shared equally: Slide 5/18

Metal-N 2 Complexes • N 2 bonds to transition metals in a similar way to CO (see Lecture 5 Slide 5) Ø σ-donation from N≡N: M Ø p-back donation from M: N≡N: (reduces N≡N bond strength) Ø synergic Ø weaker than for M-CO free N 2: v. N≡N = 2331 cm-1 1900– 2200 cm-1 ~2100 cm-1 Slide 6/18

Fixing Nitrogen • Nature needs to obtain nitrogen from atmospheric N 2 Ø nitrogen content of soil is often the growth limiting factor Ø around 17 million tonnes of NH 3 per year industrially Ø natural fixation about 2. 5 times more important • To use atmospheric N 2, nature uses metal-enzymes: Ø nitrogen fixation (nitrogenase) converts N 2 to NH 4+ Ø nitrification converts NH 4+ to NO 3 - which can be taken up by plants • Nitrogenase faces a number of problems: Ø thermodynamic stability of N≡N bond means reaction is unfavourable Ø N 2 is non-polar and unreactive: efficient enzymes/catalysts are required Ø M-N 2 bonds are weak Slide 7/18

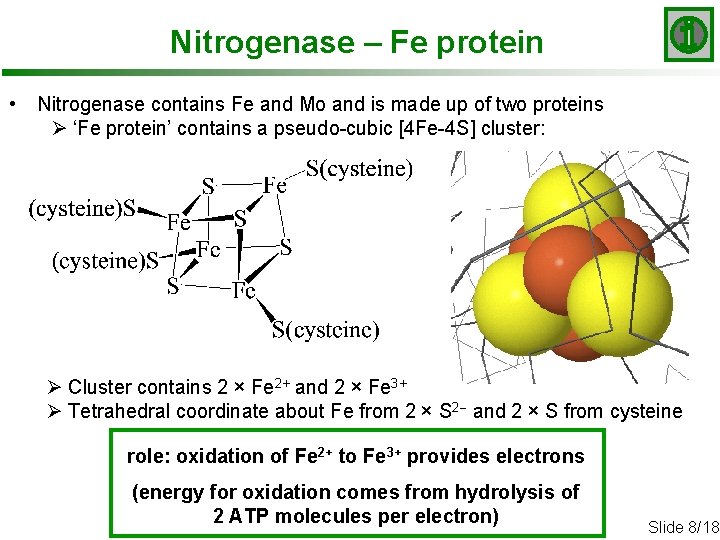
Nitrogenase – Fe protein • Nitrogenase contains Fe and Mo and is made up of two proteins Ø ‘Fe protein’ contains a pseudo-cubic [4 Fe-4 S] cluster: Ø Cluster contains 2 × Fe 2+ and 2 × Fe 3+ Ø Tetrahedral coordinate about Fe from 2 × S 2 - and 2 × S from cysteine role: oxidation of Fe 2+ to Fe 3+ provides electrons (energy for oxidation comes from hydrolysis of 2 ATP molecules per electron) Slide 8/18

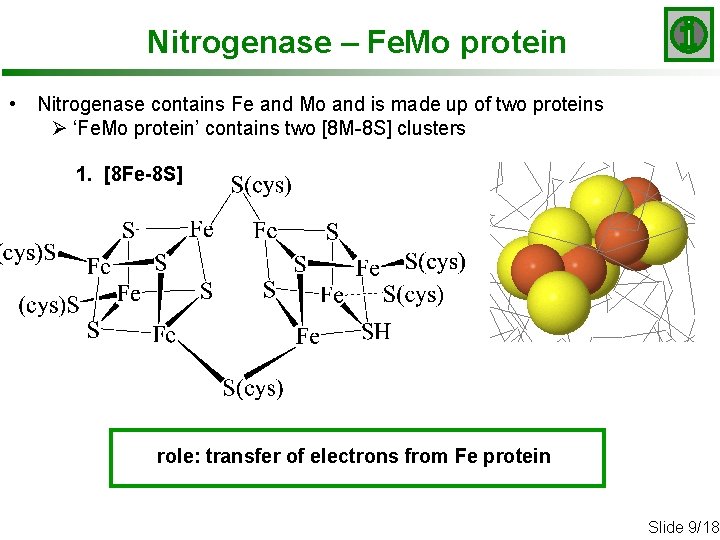
Nitrogenase – Fe. Mo protein • Nitrogenase contains Fe and Mo and is made up of two proteins Ø ‘Fe. Mo protein’ contains two [8 M-8 S] clusters 1. [8 Fe-8 S] role: transfer of electrons from Fe protein Slide 9/18

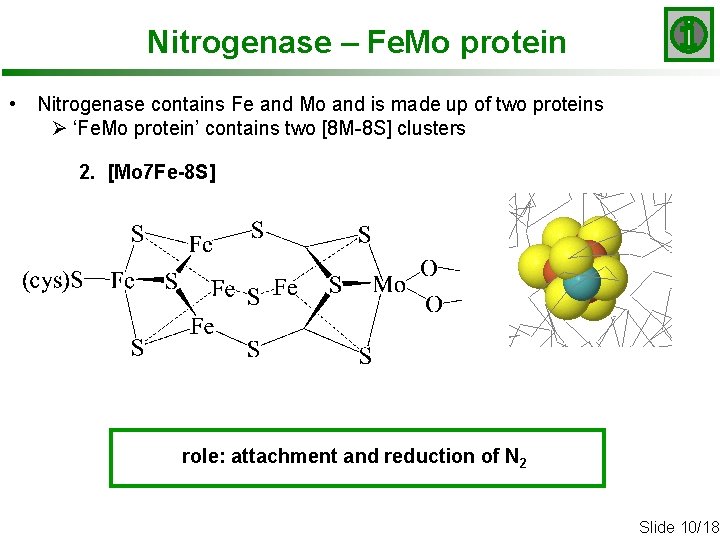
Nitrogenase – Fe. Mo protein • Nitrogenase contains Fe and Mo and is made up of two proteins Ø ‘Fe. Mo protein’ contains two [8 M-8 S] clusters 2. [Mo 7 Fe-8 S] role: attachment and reduction of N 2 Slide 10/18

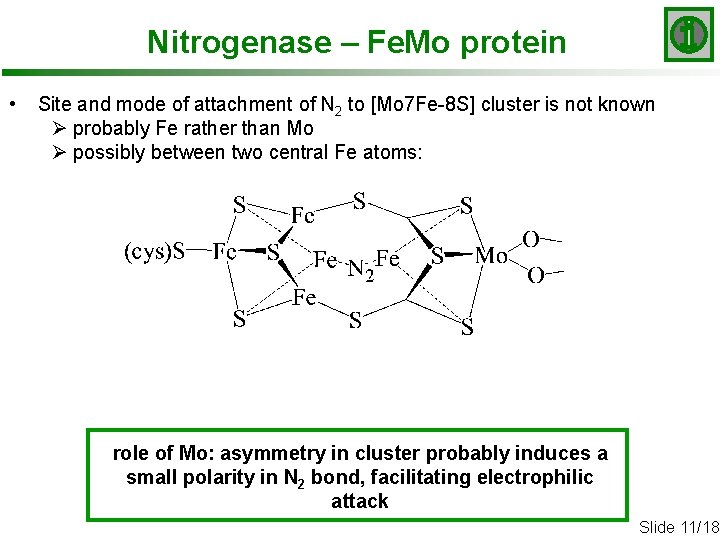
Nitrogenase – Fe. Mo protein • Site and mode of attachment of N 2 to [Mo 7 Fe-8 S] cluster is not known Ø probably Fe rather than Mo Ø possibly between two central Fe atoms: role of Mo: asymmetry in cluster probably induces a small polarity in N 2 bond, facilitating electrophilic attack Slide 11/18

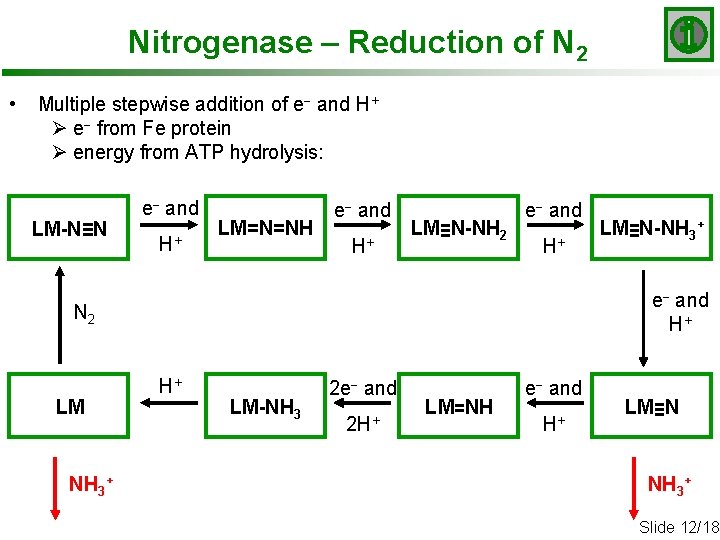
Nitrogenase – Reduction of N 2 • Multiple stepwise addition of e- and H+ Ø e- from Fe protein Ø energy from ATP hydrolysis: LM-N≡N e- and H+ LM=N=NH e- and H+ LM≡N-NH 2 e- and H+ N 2 LM NH 3+ LM≡N-NH 3+ H+ LM-NH 3 2 e- and 2 H+ LM=NH e- and H+ LM≡N NH 3+ Slide 12/18

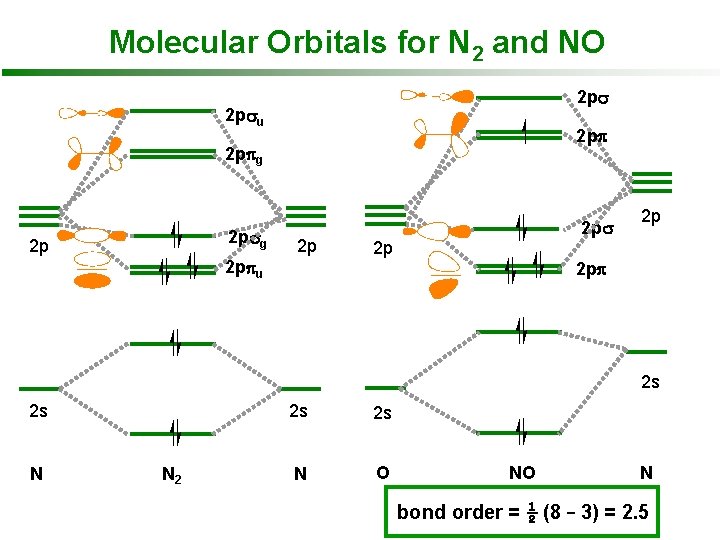
Molecular Orbitals for N 2 and NO 2 psu 2 ppg 2 psg 2 p 2 p 2 ppu 2 ps 2 p 2 p 2 pp 2 s 2 s N N 2 2 s 2 s N O NO N bond order = ½ (8 – 3) = 2. 5

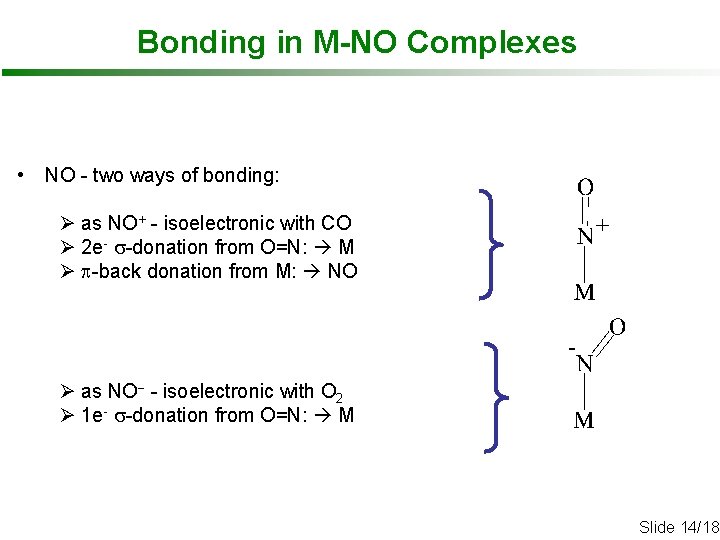
Bonding in M-NO Complexes • NO - two ways of bonding: Ø as NO+ - isoelectronic with CO Ø 2 e- s-donation from O=N: M Ø p-back donation from M: NO Ø as NO- - isoelectronic with O 2 Ø 1 e- s-donation from O=N: M Slide 14/18

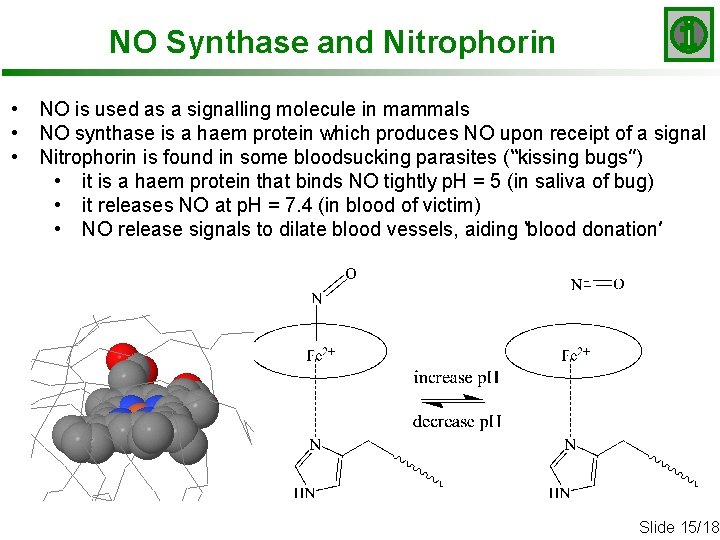
NO Synthase and Nitrophorin • • • NO is used as a signalling molecule in mammals NO synthase is a haem protein which produces NO upon receipt of a signal Nitrophorin is found in some bloodsucking parasites (“kissing bugs”) • it is a haem protein that binds NO tightly p. H = 5 (in saliva of bug) • it releases NO at p. H = 7. 4 (in blood of victim) • NO release signals to dilate blood vessels, aiding ‘blood donation’ Slide 15/18

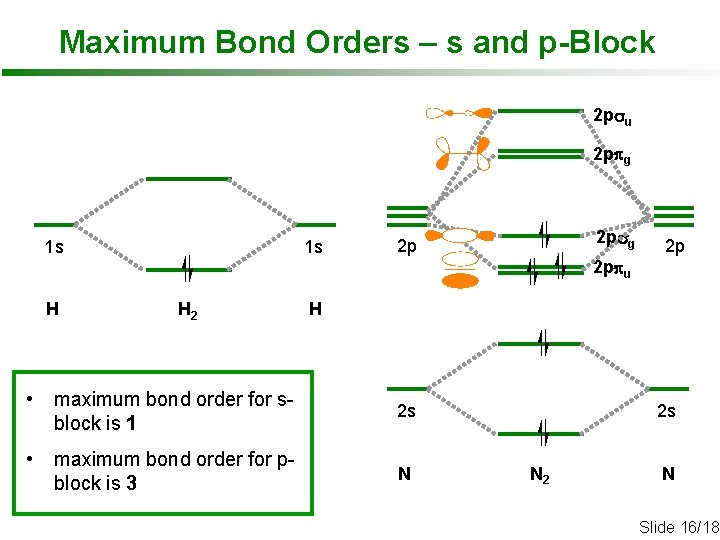
Maximum Bond Orders – s and p-Block 2 psu 2 ppg 1 s 1 s 2 psg 2 p 2 p 2 ppu H H 2 H • maximum bond order for sblock is 1 2 s • maximum bond order for pblock is 3 N 2 s N 2 N Slide 16/18

Maximum Bond Order – d-Block 3 dsu • the maximum bond order is 5 3 dpg • but…complexes also contain ligands which use some of the dorbitals reducing number of bonds 3 ddu 3 d 3 d 2× dx 2 -y 2 + dx 2 -y 2 dxy + dxy 2× dxz + dxz dyz + dyz 3 ddg 3 dpu 3 dsg dz 2 + d z 2 Slide 17/18

Summary By now you should be able to. . • Explain that why M-N 2 bonding is much weaker than MCO bonding • Count electrons in NO complexes containing bent M-NO (1 e- donor) or conraining linear M-NO (2 e- donor Next lecture • Metal-Metal bonding in complexes Slide 18/18