Sample preparation Protein and peptide separation techniques Karel



- Slides: 31

Sample preparation Protein and peptide separation techniques Karel Bezstarosti (Proteomics Center, Erasmus MC)

Proteomics 1. Identify all proteins present in a sample. 2. Characterize the post-translational modifications (PTM’s)of these proteins. 3. Protein quantification.

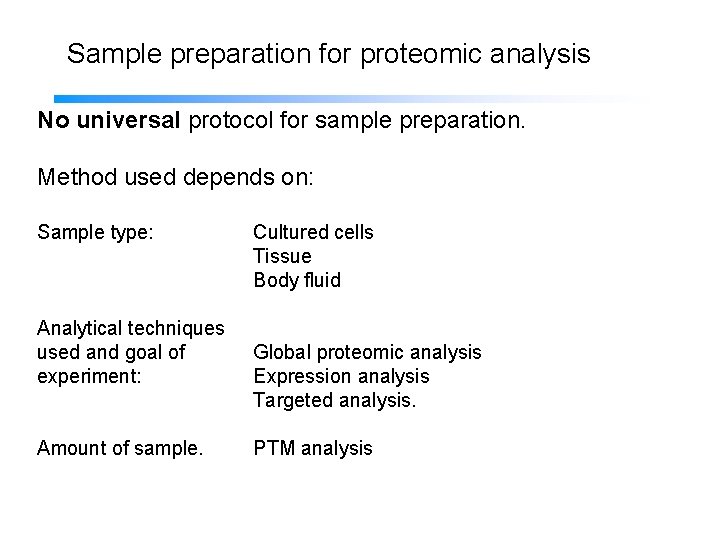
Sample preparation for proteomic analysis No universal protocol for sample preparation. Method used depends on: Sample type: Cultured cells Tissue Body fluid Analytical techniques used and goal of experiment: Global proteomic analysis Expression analysis Targeted analysis. Amount of sample. PTM analysis

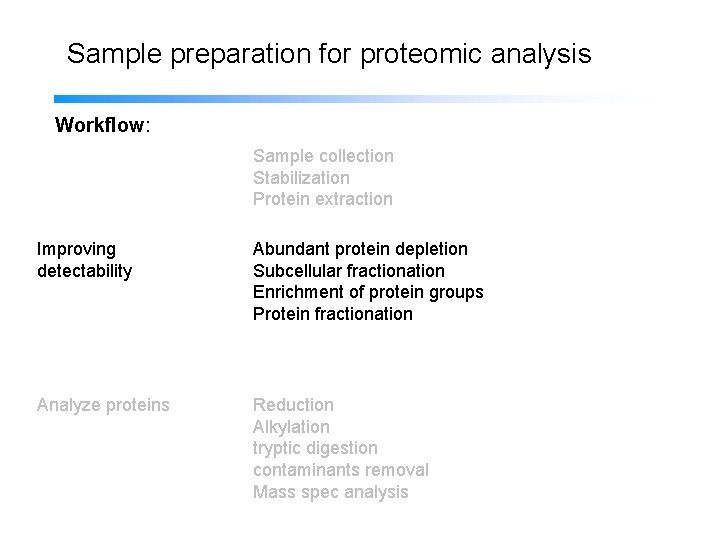
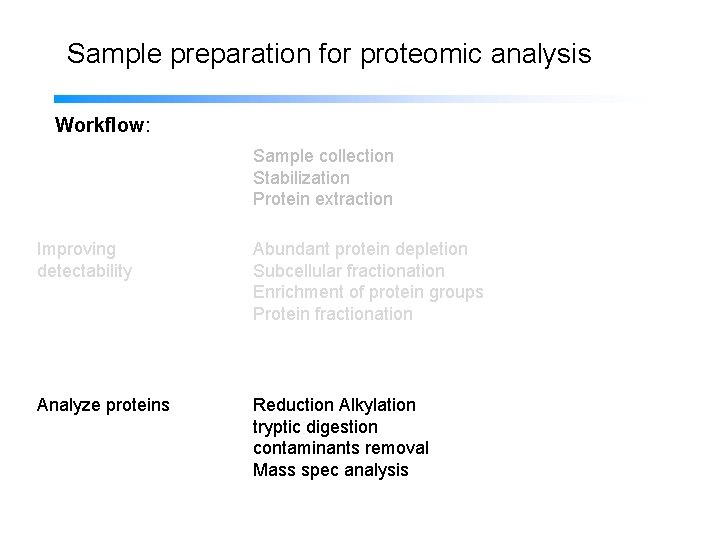
Sample preparation for proteomic analysis Workflow: Sample collection Stabilization Protein extraction Improving detectability Subcellular fractionation Abundant protein depletion Enrichment of protein groups Protein fractionation Analyze proteins Reduction Alkylation tryptic digestion contaminants removal Mass spec analysis

Sample preparation for proteomic analysis Sample collection and stabilization: Protect proteins from degradation and modification Cultured cells: During sample collection keep sample as cold as possible. Use protease and phosphatase inhibitors in solutions Inactivate enzym activity by heat (>80 OC) Tissue: Snap freeze sample in liquid nitrogen. Protect protein sample from contamination(Keratins, Detergent ……) Wear gloves when handling samples

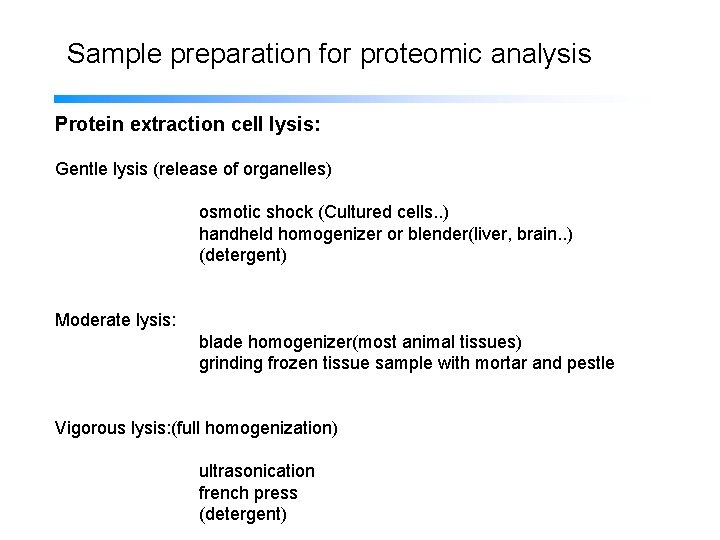
Sample preparation for proteomic analysis Protein extraction cell lysis: Gentle lysis (release of organelles) osmotic shock (Cultured cells. . ) handheld homogenizer or blender(liver, brain. . ) (detergent) Moderate lysis: blade homogenizer(most animal tissues) grinding frozen tissue sample with mortar and pestle Vigorous lysis: (full homogenization) ultrasonication french press (detergent)

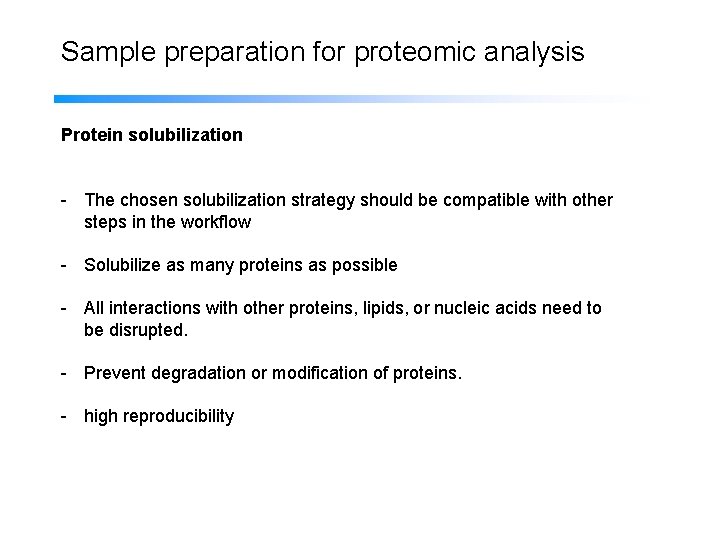
Sample preparation for proteomic analysis Protein solubilization - The chosen solubilization strategy should be compatible with other steps in the workflow - Solubilize as many proteins as possible - All interactions with other proteins, lipids, or nucleic acids need to be disrupted. - Prevent degradation or modification of proteins. - high reproducibility

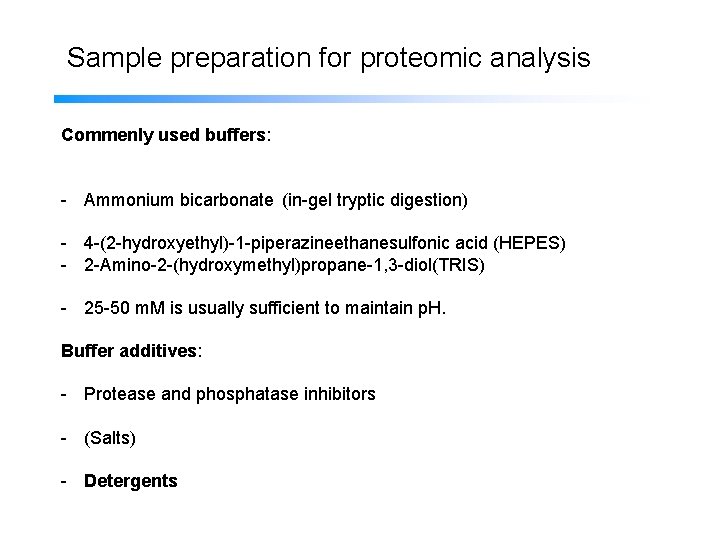
Sample preparation for proteomic analysis Commenly used buffers: - Ammonium bicarbonate (in-gel tryptic digestion) - 4 -(2 -hydroxyethyl)-1 -piperazineethanesulfonic acid (HEPES) - 2 -Amino-2 -(hydroxymethyl)propane-1, 3 -diol(TRIS) - 25 -50 m. M is usually sufficient to maintain p. H. Buffer additives: - Protease and phosphatase inhibitors - (Salts) - Detergents

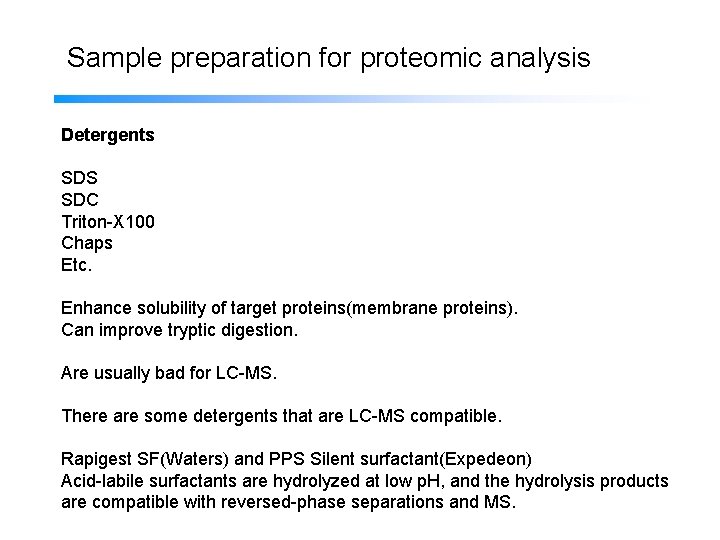
Sample preparation for proteomic analysis Detergents SDS SDC Triton-X 100 Chaps Etc. Enhance solubility of target proteins(membrane proteins). Can improve tryptic digestion. Are usually bad for LC-MS. There are some detergents that are LC-MS compatible. Rapigest SF(Waters) and PPS Silent surfactant(Expedeon) Acid-labile surfactants are hydrolyzed at low p. H, and the hydrolysis products are compatible with reversed-phase separations and MS.

Sample preparation for proteomic analysis Workflow: Sample collection Stabilization Protein extraction Improving detectability Abundant protein depletion Subcellular fractionation Enrichment of protein groups Protein fractionation Analyze proteins Reduction Alkylation tryptic digestion contaminants removal Mass spec analysis

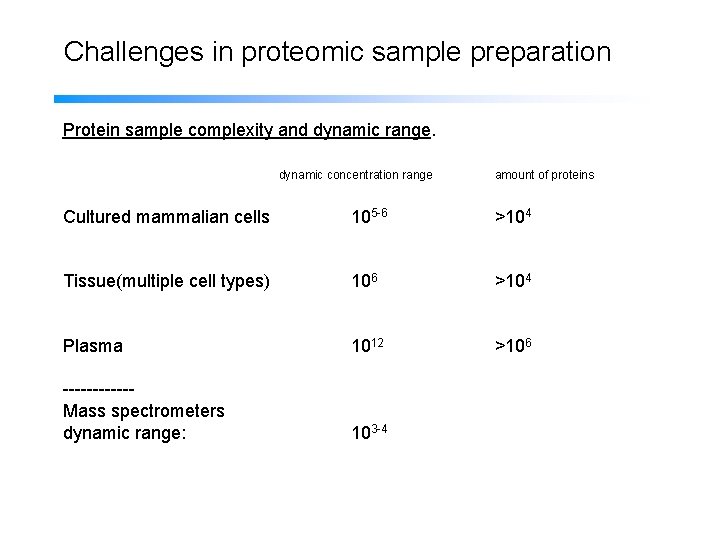
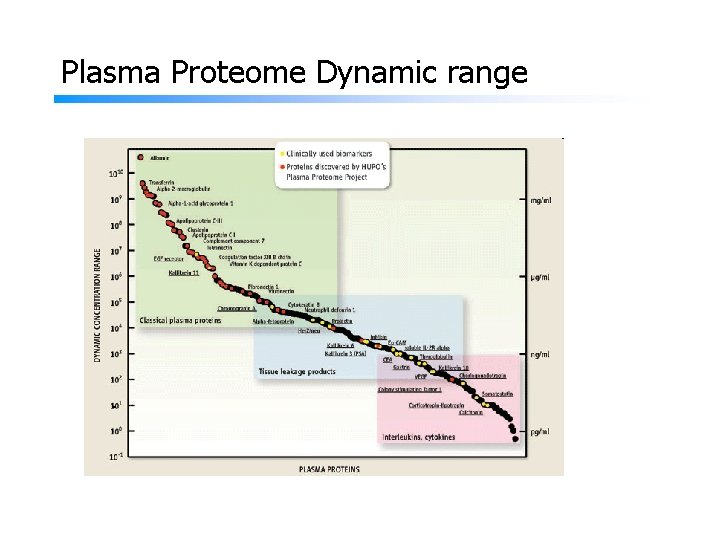
Challenges in proteomic sample preparation Protein sample complexity and dynamic range. dynamic concentration range amount of proteins Cultured mammalian cells 105 -6 >104 Tissue(multiple cell types) 106 >104 Plasma 1012 >106 ------Mass spectrometers dynamic range: 103 -4

Plasma Proteome Dynamic range

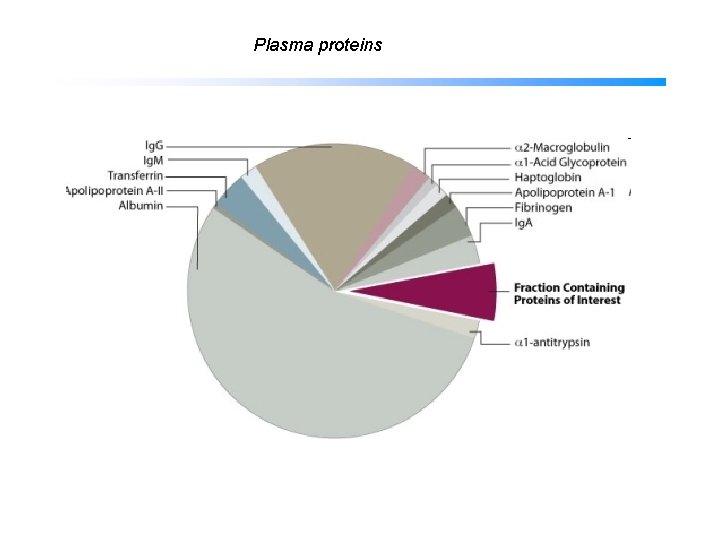
Plasma proteins

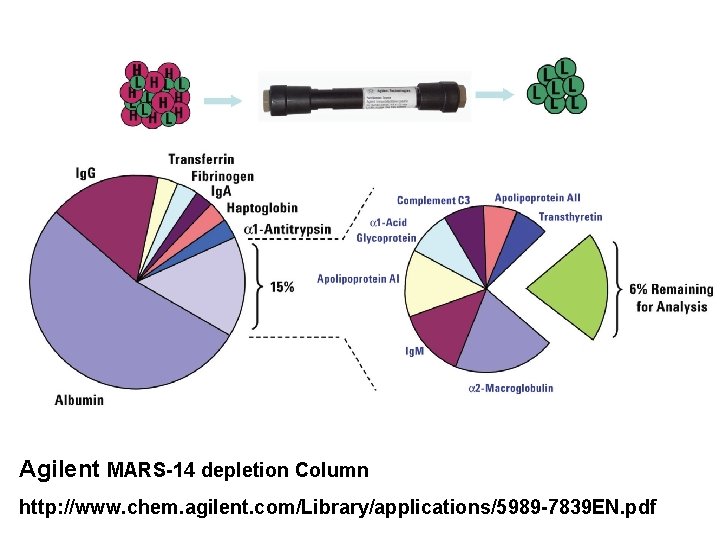
Depletion of Abundant Plasma Proteins Agilent MARS-14 depletion Column http: //www. chem. agilent. com/Library/applications/5989 -7839 EN. pdf

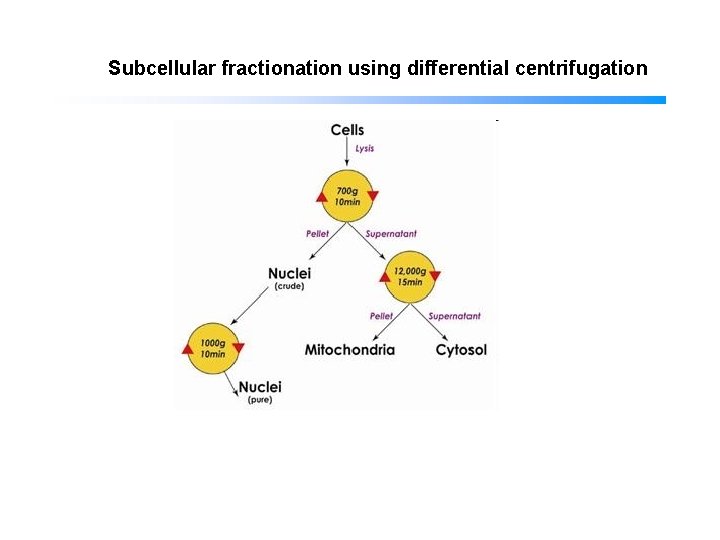
Subcellular fractionation using differential centrifugation

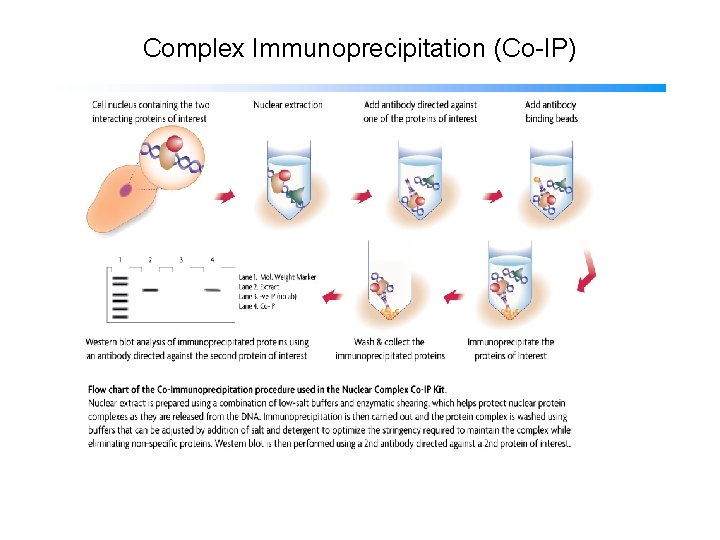
Complex Immunoprecipitation (Co-IP)

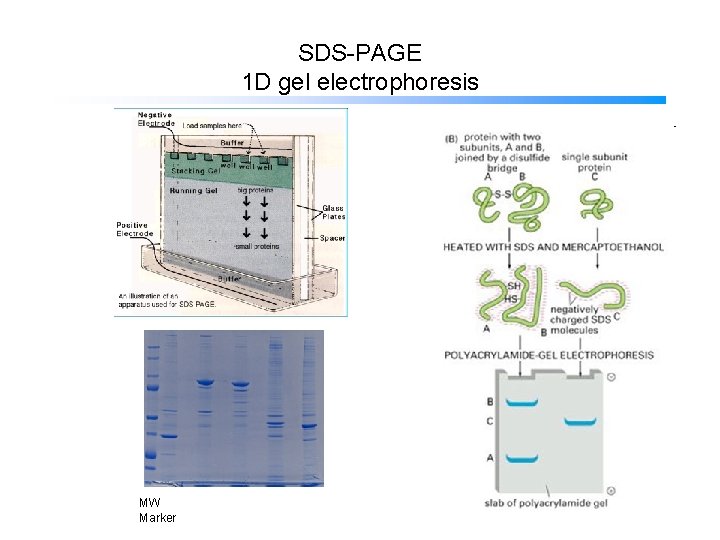
SDS-PAGE 1 D gel electrophoresis MW Marker

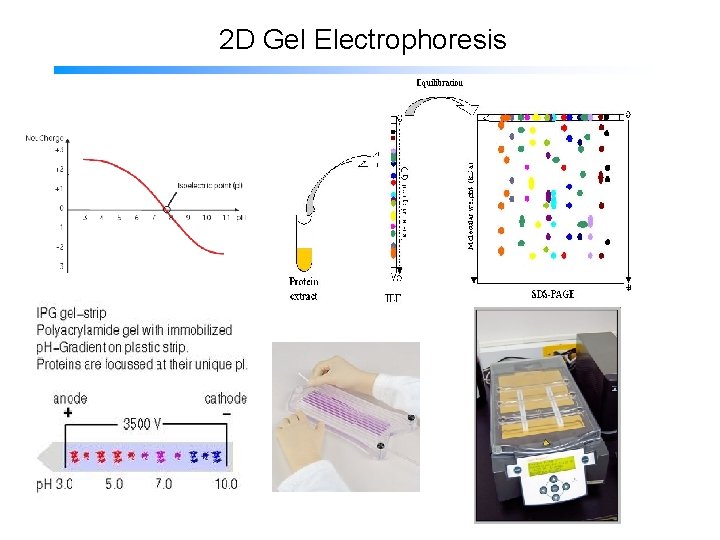
2 D Gel Electrophoresis

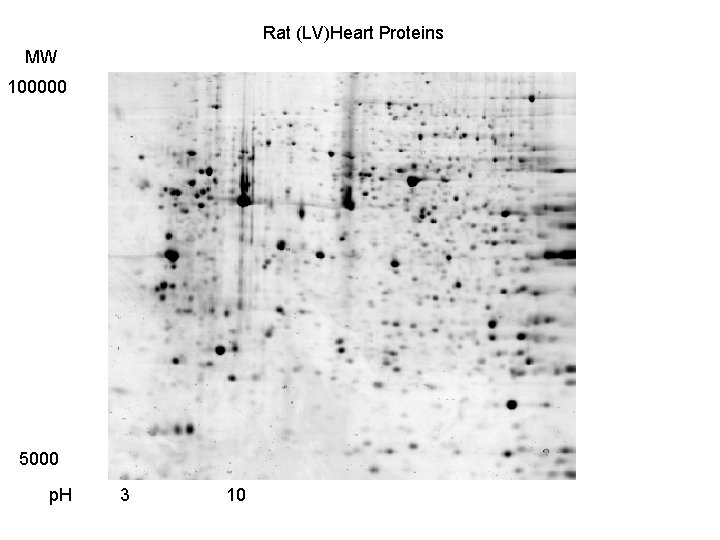
Rat (LV)Heart Proteins MW 100000 5000 p. H 3 10

Sample preparation for proteomic analysis Workflow: Sample collection Stabilization Protein extraction Improving detectability Abundant protein depletion Subcellular fractionation Enrichment of protein groups Protein fractionation Analyze proteins Reduction Alkylation tryptic digestion contaminants removal Mass spec analysis

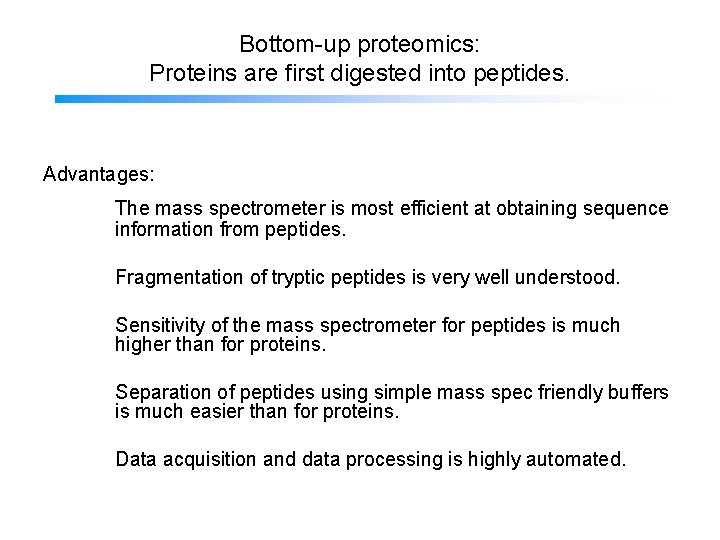
Bottom-up proteomics: Proteins are first digested into peptides. Advantages: The mass spectrometer is most efficient at obtaining sequence information from peptides. Fragmentation of tryptic peptides is very well understood. Sensitivity of the mass spectrometer for peptides is much higher than for proteins. Separation of peptides using simple mass spec friendly buffers is much easier than for proteins. Data acquisition and data processing is highly automated.

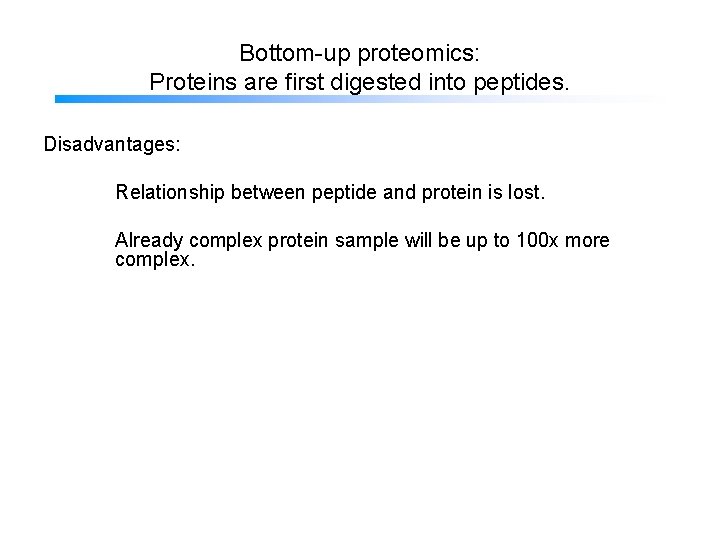
Bottom-up proteomics: Proteins are first digested into peptides. Disadvantages: Relationship between peptide and protein is lost. Already complex protein sample will be up to 100 x more complex.

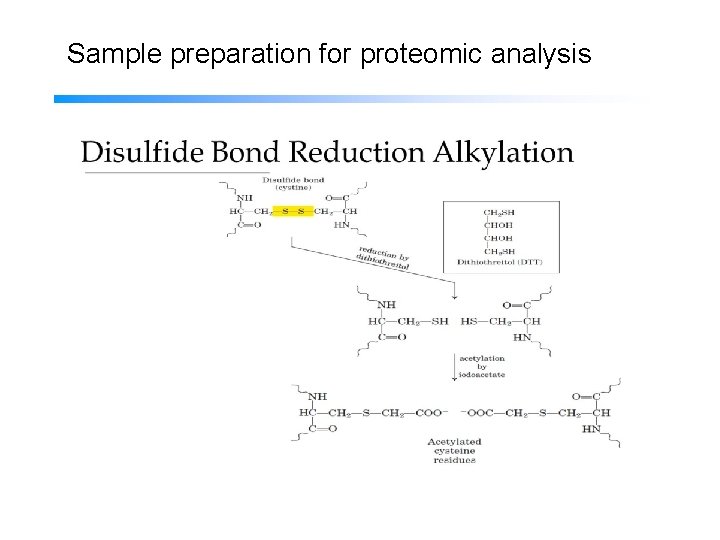
Sample preparation for proteomic analysis

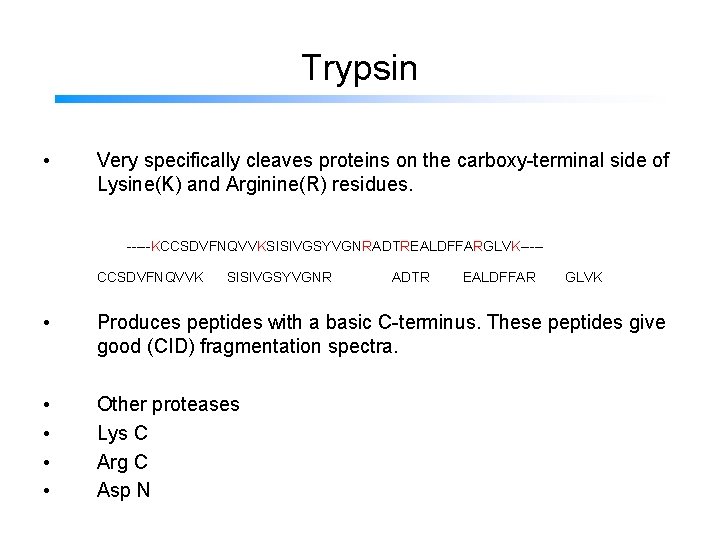
Trypsin • Very specifically cleaves proteins on the carboxy-terminal side of Lysine(K) and Arginine(R) residues. -----KCCSDVFNQVVKSISIVGSYVGNRADTREALDFFARGLVK----CCSDVFNQVVK SISIVGSYVGNR ADTR EALDFFAR GLVK • Produces peptides with a basic C-terminus. These peptides give good (CID) fragmentation spectra. • • Other proteases Lys C Arg C Asp N

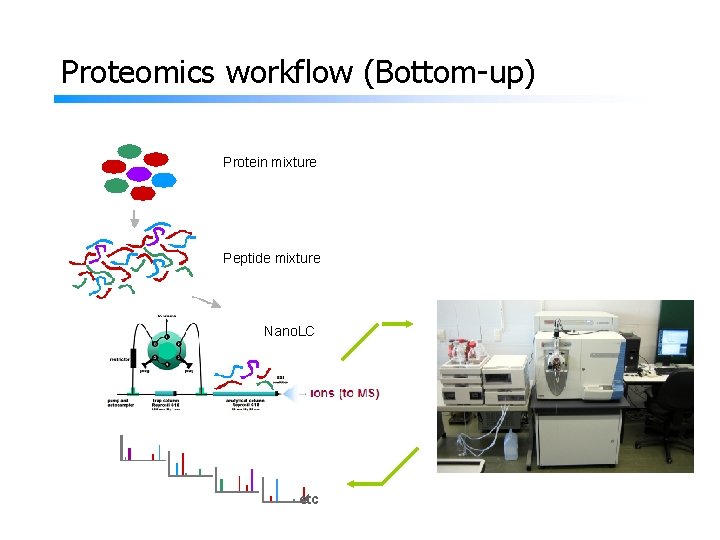
Proteomics workflow (Bottom-up) Protein mixture Peptide mixture Nano. LC etc

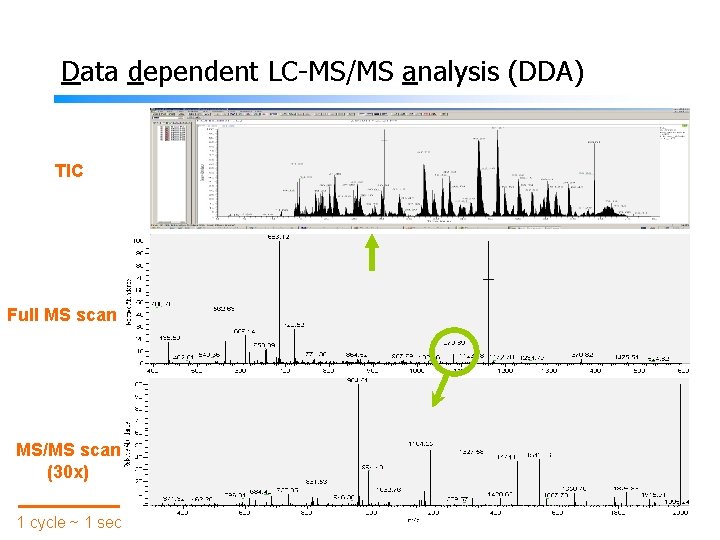
Data dependent LC-MS/MS analysis (DDA) TIC Full MS scan MS/MS scan (30 x) 1 cycle ~ 1 sec

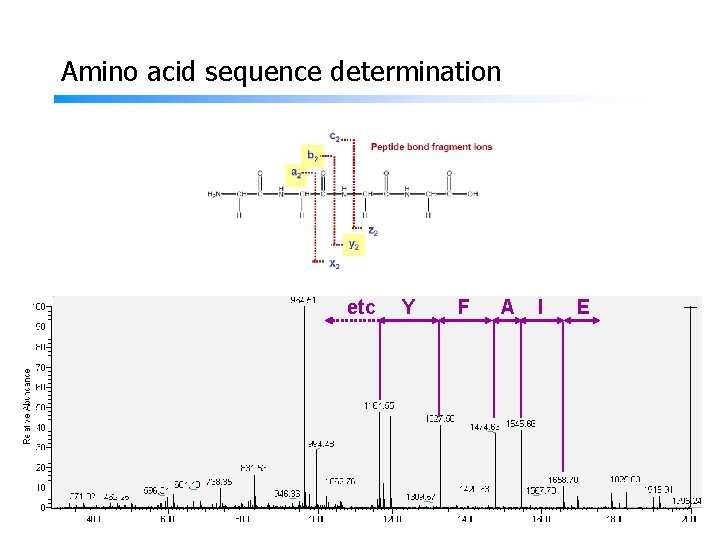
Amino acid sequence determination etc Y F A I E

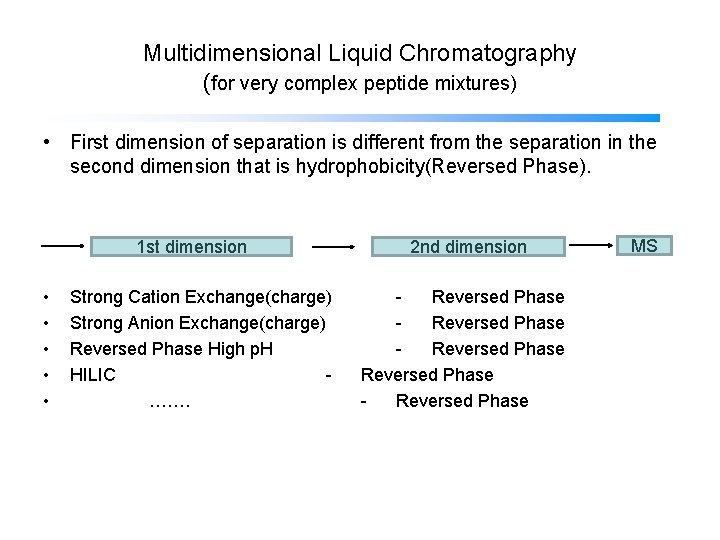
Multidimensional Liquid Chromatography (for very complex peptide mixtures) • First dimension of separation is different from the separation in the second dimension that is hydrophobicity(Reversed Phase). 1 st dimension • • • Strong Cation Exchange(charge) Strong Anion Exchange(charge) Reversed Phase High p. H HILIC ……. 2 nd dimension Reversed Phase Reversed Phase MS

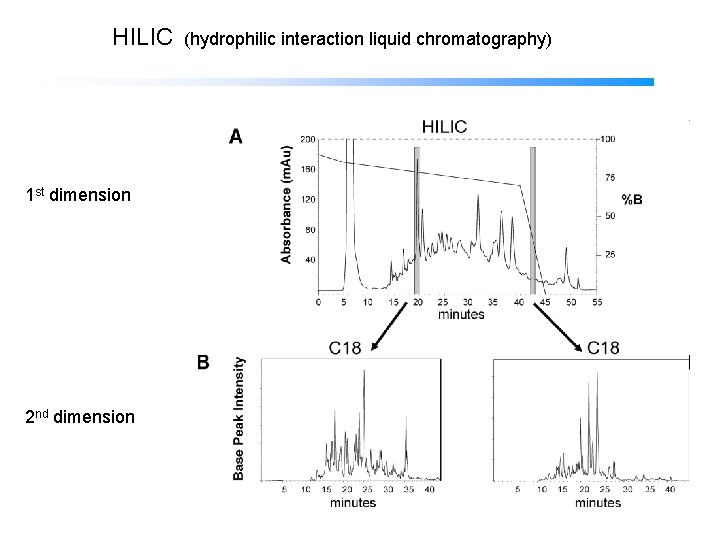
HILIC (hydrophilic interaction liquid chromatography) 1 st dimension 2 nd dimension

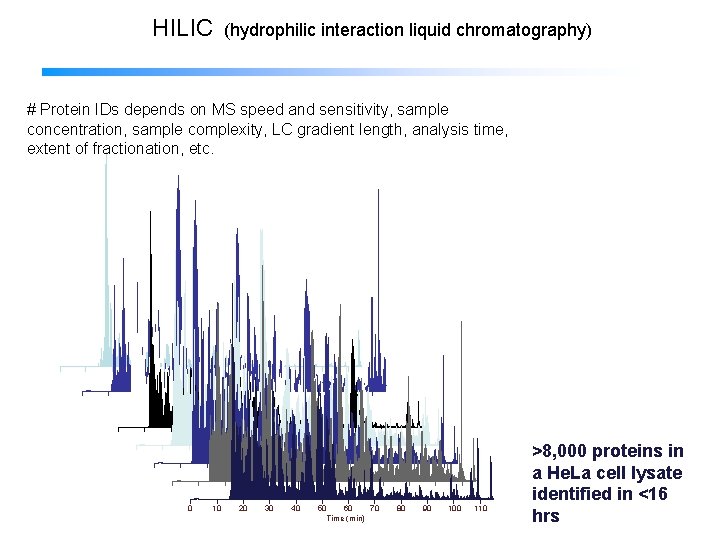
HILIC (hydrophilic interaction liquid chromatography) HILIC fractionation and LC-MS # Protein IDs depends on MS speed and sensitivity, sample concentration, sample complexity, LC gradient length, analysis time, extent of fractionation, etc. 0 10 20 30 40 50 60 70 Time (min) 80 90 100 110 >8, 000 proteins in a He. La cell lysate identified in <16 hrs

Please feel free to contact us so that we can help you design your experiments. http: //www. proteomicscenter. nl/ Thank you Questions?