Rowland NMR Toolkit RNMRTK Overview Originally developed as



- Slides: 17

Rowland NMR Toolkit (RNMRTK) Overview • Originally developed as a platform for developing new NMR data processing methods • Used primarily for the purposes of running Max. Ent • Provides • Rich set of apodization (window) functions • DFT processing • General processing tools (phasing, reversing, truncating, etc. ) • Robust, efficient Linear Prediction (LP) extrapolation • Max. Ent reconstruction (deconvolution, uniform or nonuniform sampling) • Efficient parallel processing • exports to nmr. Pipe, XEASY, Felix; NMRView. J reads RNMRTK format. • Generates synthetic data, noise (testing/error analysis) • Free for Academic use and included in NMRbox • l 1 -norm real (LONER) has recently been added

RNMRTK – Suite of Programs section – Manages the shared memory sections rnmrtk – The main processing program in the Toolkit Loading, apodization, DFT, phasing, saving, etc. flip – Forward linear prediction msa / msa 2 d / msa 3 d – Maximum entropy reconstruction in one, two, and three dimensions inject – Add synthetic peaks to time domain data select / zsample – Expands or compresses NUS data seepln / contour – graphical display for 1 D and 2 D data Many others …

RNMRTK – Shared memory With a suite of programs for performing tasks – How do you get data output from one command to the next command without needing to write out intermediate files? • Pipes (nmr. Pipe strategy) • Shared memory (RNMRTK strategy) • A shared memory section is a chunk of persistent memory where NMR data can be stored and used by different programs. • /dev/shm – A modern version of a shared memory implementation that gives the performance of shared memory, but without the overhead of managing traditional shared memory section • RNMRTK program to manage (create / delete) shared memory sections section -c 1024 96 45 • The size of the shared memory section is the product of the arguments multiplied by 4 (32 bit data) plus 512 extra bytes for a header. • Shared memory sections must be large enough to store all the data – including processing section -d • Deletes the shared memory section

RNMRTK – Shared memory The maximum size of shared memory sections is defined by kernel values shmmax and shmall. NMRbox takes care of these advanced settings. Example: Assume a data set size of 512 x 128 complex points “section -c 512 128” Shared memory section is too small as data is complex “section -c 1024 256” Shared memory section just fits data, but cannot handled a ZF “section -c 2048 512” Shared memory section allows each dimension to be doubled NOTES: “section -d” Shared memory sections stay persistent until forcefully deleted or the computer rebooted “section -c 1024” Shared memory sections will delete exisiting shared memory sections when created No harm in having shared memory sections be too large

RNMRTK – rnmrtk • Command for performing many of the traditional processing techniques • Row (column) oriented. • Many commands need to have the dimension set • Can be run: • one command at a time • in interactive mode • scripted

RNMRTK – Issuing commands • Arguments are entered as different classes: • floats (defined as a number with a decimal) • setpar SF 1 599. 8763 • integers (defined as a number without a decimal) • zerofill 2048 • string (defined as a character string without a period) • sinebell square 70. 0 • filename (defined as a character string with a decimal point) • loadvnmr. /fid • load hsqc. sec • The order of arguments entered is important, but only within a given class of argument • sstdc 16 20 156. 25 COS • sstdc 156. 25 16 COS 20 • rnmrtk is case insensitive (except for filenames)

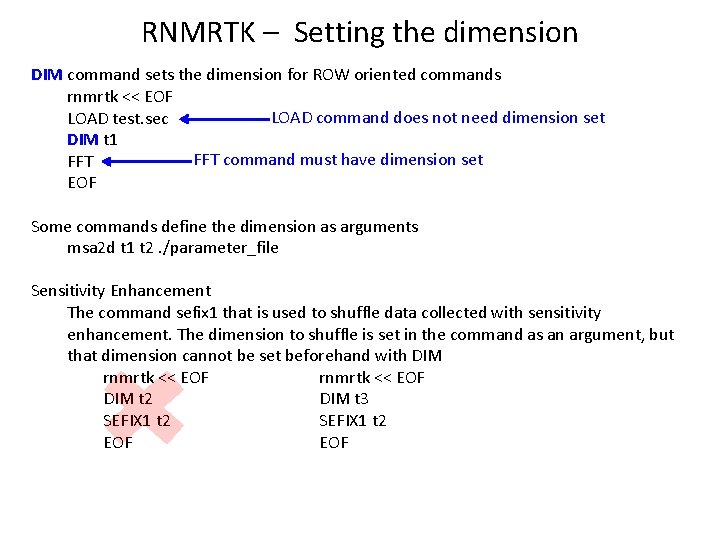
RNMRTK – Setting the dimension DIM command sets the dimension for ROW oriented commands rnmrtk << EOF LOAD command does not need dimension set LOAD test. sec DIM t 1 FFT command must have dimension set FFT EOF Some commands define the dimension as arguments msa 2 d t 1 t 2. /parameter_file Sensitivity Enhancement The command sefix 1 that is used to shuffle data collected with sensitivity enhancement. The dimension to shuffle is set in the command as an argument, but that dimension cannot be set beforehand with DIM rnmrtk << EOF DIM t 2 DIM t 3 SEFIX 1 t 2 EOF

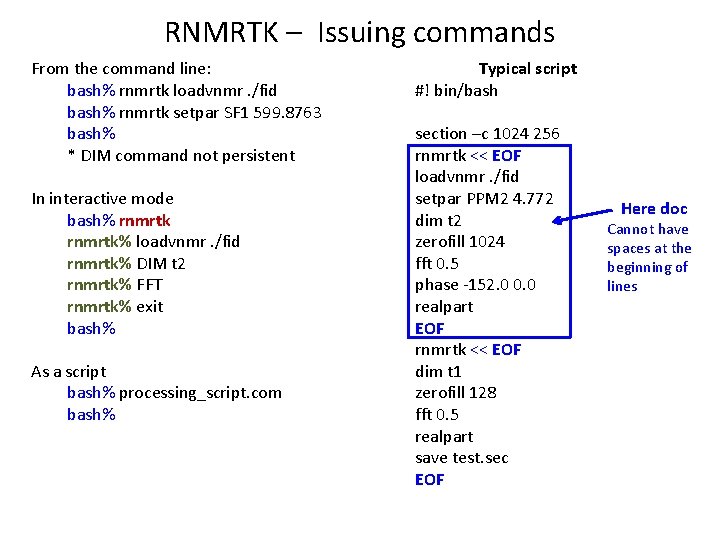
RNMRTK – Issuing commands From the command line: bash% rnmrtk loadvnmr. /fid bash% rnmrtk setpar SF 1 599. 8763 bash% * DIM command not persistent In interactive mode bash% rnmrtk% loadvnmr. /fid rnmrtk% DIM t 2 rnmrtk% FFT rnmrtk% exit bash% As a script bash% processing_script. com bash% Typical script #! bin/bash section –c 1024 256 rnmrtk << EOF loadvnmr. /fid setpar PPM 2 4. 772 dim t 2 zerofill 1024 fft 0. 5 phase -152. 0 0. 0 realpart EOF rnmrtk << EOF dim t 1 zerofill 128 fft 0. 5 realpart save test. sec EOF Here doc Cannot have spaces at the beginning of lines

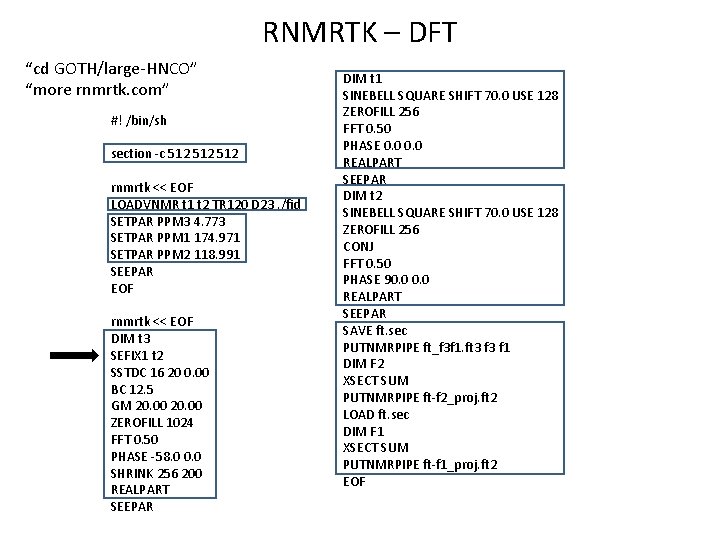
RNMRTK – DFT “cd GOTH/large-HNCO” “more rnmrtk. com” #! /bin/sh section -c 512 512 rnmrtk << EOF LOADVNMR t 1 t 2 TR 120 D 23. /fid SETPAR PPM 3 4. 773 SETPAR PPM 1 174. 971 SETPAR PPM 2 118. 991 SEEPAR EOF rnmrtk << EOF DIM t 3 SEFIX 1 t 2 SSTDC 16 20 0. 00 BC 12. 5 GM 20. 00 ZEROFILL 1024 FFT 0. 50 PHASE -58. 0 0. 0 SHRINK 256 200 REALPART SEEPAR DIM t 1 SINEBELL SQUARE SHIFT 70. 0 USE 128 ZEROFILL 256 FFT 0. 50 PHASE 0. 0 REALPART SEEPAR DIM t 2 SINEBELL SQUARE SHIFT 70. 0 USE 128 ZEROFILL 256 CONJ FFT 0. 50 PHASE 90. 0 REALPART SEEPAR SAVE ft. sec PUTNMRPIPE ft_f 3 f 1. ft 3 f 1 DIM F 2 XSECT SUM PUTNMRPIPE ft-f 2_proj. ft 2 LOAD ft. sec DIM F 1 XSECT SUM PUTNMRPIPE ft-f 1_proj. ft 2 EOF

Maximum Entropy Reconstruction (Max. Ent) • Max. Ent can be performed in one dimension (msa), two dimensions (msa 2 d) and three dimensions (msa 4 d) – NOTE: # of dimension Max. Ent is acting on, not dimension of experiment. • The acquisition dimension is often processed with a DFT and not with Max. Ent • There are two modes for processing with Max. Ent • Constant AIM • Must define DEF and AIM • Constant LAMBDA • Must define DEF and LAMBDA • When all the dimensions are NOT processed with Max. Ent, such as when the acquisition dimension is processed with a DFT, you MUST use Constant Lambda mode • 2 D Data sets • Option 1: Process F 2 with DFT, and F 1 with msa (lambda mode) • Option 2: Process F 2 -F 1 planes with msa 2 d (aim mode) • 3 D Data sets • Option 1: Process F 3 with DFT and F 1 -F 2 planes with msa 2 d (lambda mode) • Option 2: Process F 3 -F 2 -F 1 cube with msa 3 d (aim mode) • 4 D Data sets • Option 1: Process F 4 with DFT and F 1 -F 2 -F 3 cubes with msa 3 d (lambda mode)

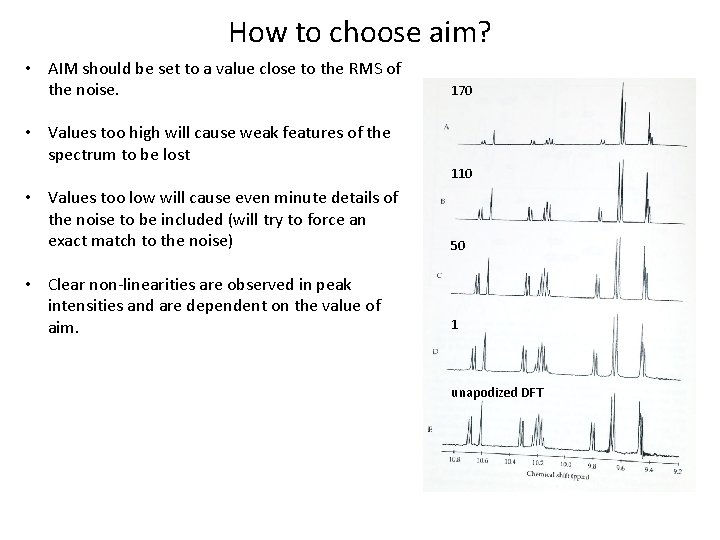
How to choose aim? • AIM should be set to a value close to the RMS of the noise. 170 • Values too high will cause weak features of the spectrum to be lost 110 • Values too low will cause even minute details of the noise to be included (will try to force an exact match to the noise) 50 • Clear non-linearities are observed in peak intensities and are dependent on the value of aim. 1 unapodized DFT

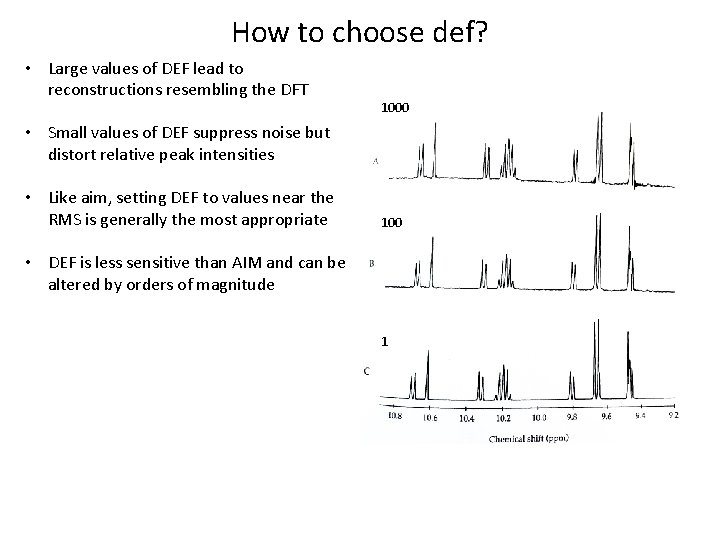
How to choose def? • Large values of DEF lead to reconstructions resembling the DFT 1000 • Small values of DEF suppress noise but distort relative peak intensities • Like aim, setting DEF to values near the RMS is generally the most appropriate 100 • DEF is less sensitive than AIM and can be altered by orders of magnitude 1

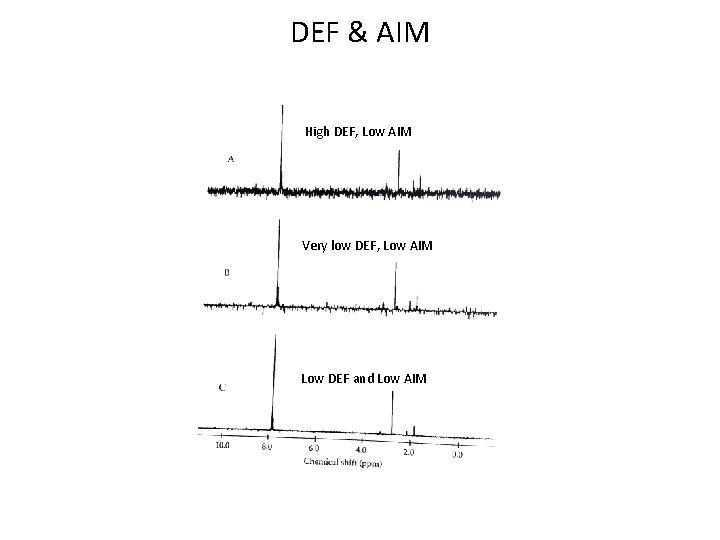
DEF & AIM High DEF, Low AIM Very low DEF, Low AIM Low DEF and Low AIM

LAMBDA • During a constant AIM calculation a value LAMBDA will be converged to. • LAMBDA is the weight applied to the entropy function relative to the constraint of matching to the experimental data • If processing 1 D slices of a 2 D; 2 D planes of a 3 D; or 3 D cubes of a 4 D, each calculation will converge to a different LAMBDA. • LAMBDA will affect the non-linearity and cause a “scaling” of the data • Let’s assume we are processing a 3 D data set where the acquisition dimension is processed with a DFT and the 2 D indirect planes are processed with Max. Ent • In constant AIM mode each plane will have a slightly different LAMBDA value and hence a slight different in scaling and non-linearity. This will cause distortions in the 3 D peak shapes • To “fix” this issue Max. Ent can be run in constant LAMBDA mode • To determine LAMBDA, it is typical to process the data initially in constant AIM mode and average the converged LAMBDA values.

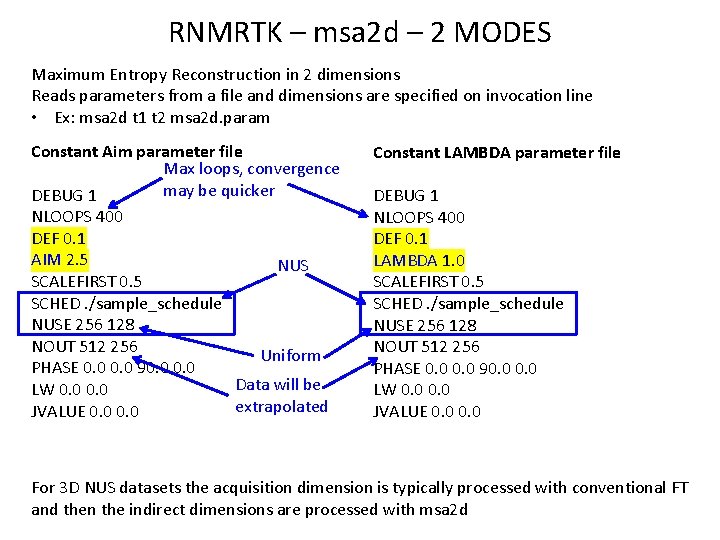
RNMRTK – msa 2 d – 2 MODES Maximum Entropy Reconstruction in 2 dimensions Reads parameters from a file and dimensions are specified on invocation line • Ex: msa 2 d t 1 t 2 msa 2 d. param Constant Aim parameter file Max loops, convergence may be quicker DEBUG 1 NLOOPS 400 DEF 0. 1 AIM 2. 5 NUS SCALEFIRST 0. 5 SCHED. /sample_schedule NUSE 256 128 NOUT 512 256 Uniform PHASE 0. 0 90. 0 Data will be LW 0. 0 extrapolated JVALUE 0. 0 Constant LAMBDA parameter file DEBUG 1 NLOOPS 400 DEF 0. 1 LAMBDA 1. 0 SCALEFIRST 0. 5 SCHED. /sample_schedule NUSE 256 128 NOUT 512 256 PHASE 0. 0 90. 0 LW 0. 0 JVALUE 0. 0 For 3 D NUS datasets the acquisition dimension is typically processed with conventional FT and then the indirect dimensions are processed with msa 2 d

Let’s try a Max. Ent Reconstruction “cd large-HNCO” “more msa 2 d_param” “wc -l schedule 3. scd” • There are 400 / 16, 384 FIDs collected (2. 4%). /msa 2 d. com schedule 3. scd test” “nmr. Draw -in test_nudft_proj. ft 2” “nmr. Draw -in test_nudft. ft 3” “nmr. Draw -in test_msa 2 d_proj. ft 2” “nmr. Draw -in test_msa 2 d. ft 3” You can compare the results to the DFT using 100% of the FIDs “nmr. Draw -in ft-f 1_proj. ft 2” “nmr. Draw -in ft-f 2_proj. ft 2” “nmr. Draw -in ft_f 3 f 1. ft 3”

RNMRTK – msa 2 d Monitoring convergence Very important! • Ensure that the value of test is less than 0. 1 unless fewer than NLOOPS required • Without deconvolution convergence typically occurs is around 40 iterations if proper DEF, AIM, and LAMBDA values were use. • High numbers of loops suggest msa 2 d is modeling noise • With deconvolution the number of loops may be significantly higher. How to check values for Test and the maximum number of loops “tail msa 2 d. txt” “grep ^Test msa 2 d. txt | sort -n -k 3” “grep Loop msa 2 d. txt | sort -n -k 2”