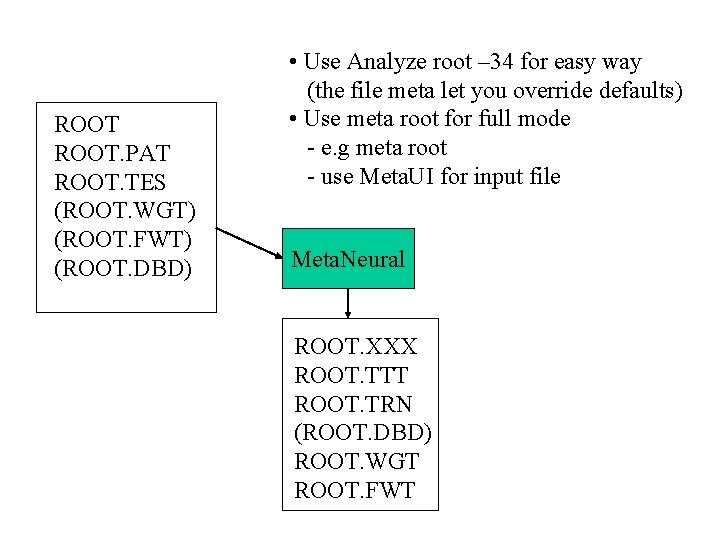
ROOT PAT ROOT TES ROOT WGT ROOT FWT


- Slides: 32

ROOT. PAT ROOT. TES (ROOT. WGT) (ROOT. FWT) (ROOT. DBD) • Use Analyze root – 34 for easy way (the file meta let you override defaults) • Use meta root for full mode - e. g meta root - use Meta. UI for input file Meta. Neural ROOT. XXX ROOT. TTT ROOT. TRN (ROOT. DBD) ROOT. WGT ROOT. FWT

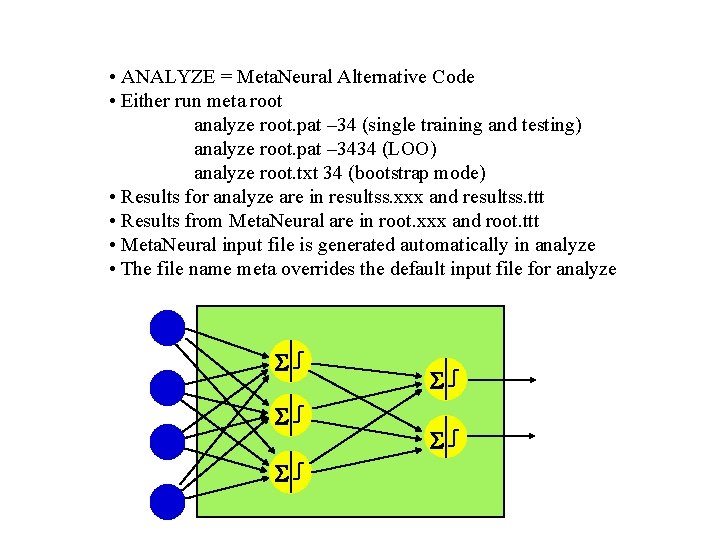
• ANALYZE = Meta. Neural Alternative Code • Either run meta root analyze root. pat – 34 (single training and testing) analyze root. pat – 3434 (LOO) analyze root. txt 34 (bootstrap mode) • Results for analyze are in resultss. xxx and resultss. ttt • Results from Meta. Neural are in root. xxx and root. ttt • Meta. Neural input file is generated automatically in analyze • The file name meta overrides the default input file for analyze S S S

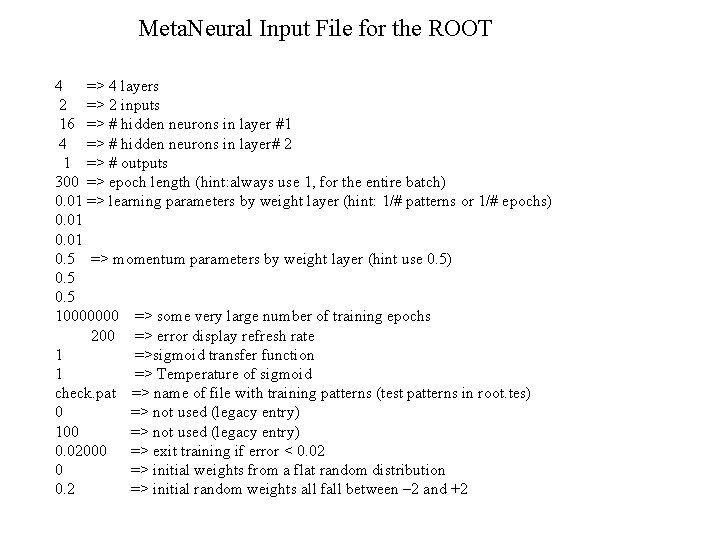
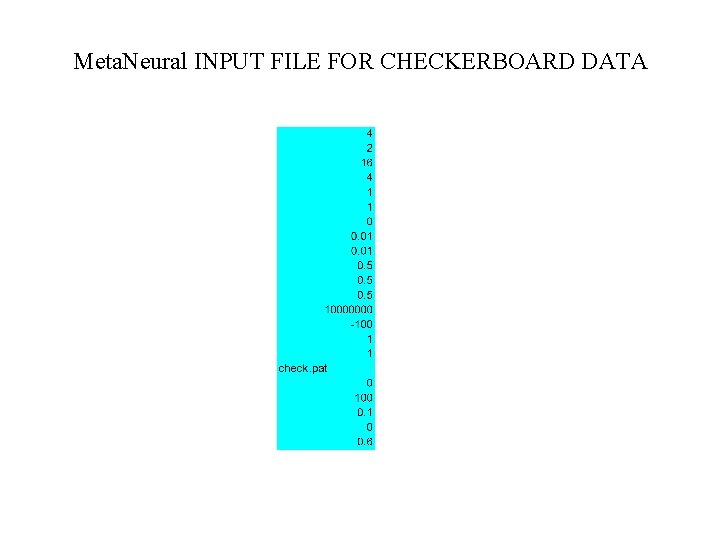
Meta. Neural Input File for the ROOT 4 => 4 layers 2 => 2 inputs 16 => # hidden neurons in layer #1 4 => # hidden neurons in layer# 2 1 => # outputs 300 => epoch length (hint: always use 1, for the entire batch) 0. 01 => learning parameters by weight layer (hint: 1/# patterns or 1/# epochs) 0. 01 0. 5 => momentum parameters by weight layer (hint use 0. 5) 0. 5 10000000 => some very large number of training epochs 200 => error display refresh rate 1 =>sigmoid transfer function 1 => Temperature of sigmoid check. pat => name of file with training patterns (test patterns in root. tes) 0 => not used (legacy entry) 100 => not used (legacy entry) 0. 02000 => exit training if error < 0. 02 0 => initial weights from a flat random distribution 0. 2 => initial random weights all fall between – 2 and +2

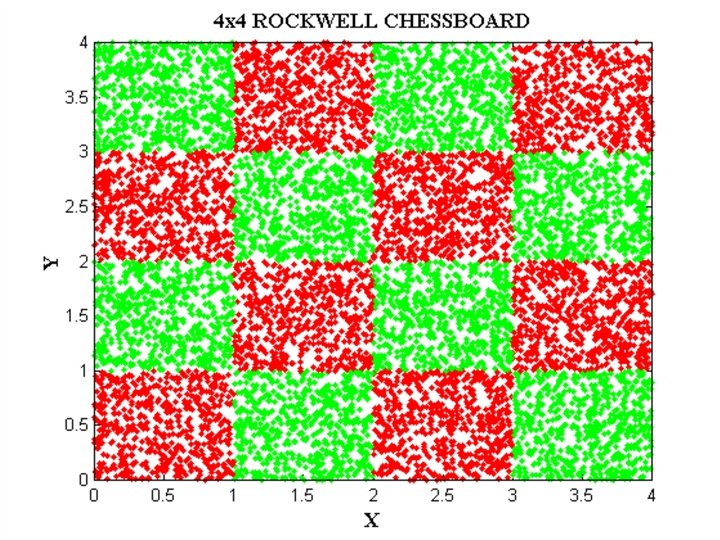
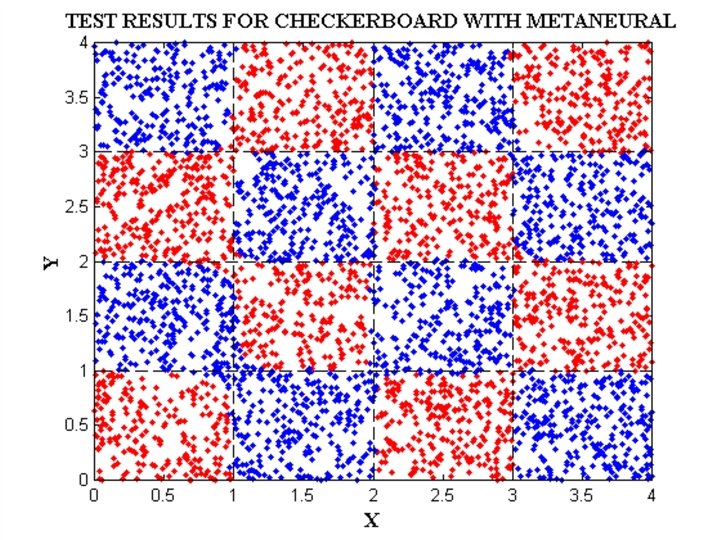
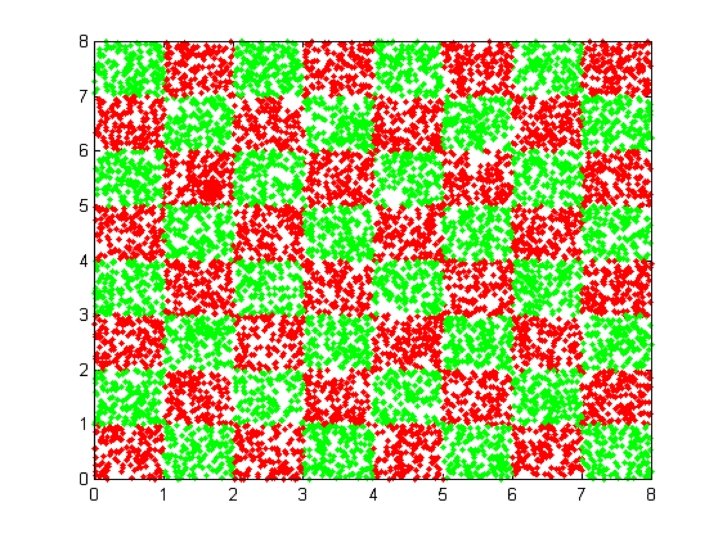
EXAMPLE DATA SETS • IRIS data • Checkerboard data • Svante wold’s QSAR data • Cherkassky’s nonlinear function • Albumin QSAR data


FILES RELATED TO CHECKERBOARD EXAMPLE CHECK_DATA. BAT CHECK_NET. BAT CHECK_TEST. BAT CHECK. PAT

Meta. Neural INPUT FILE FOR CHECKERBOARD DATA




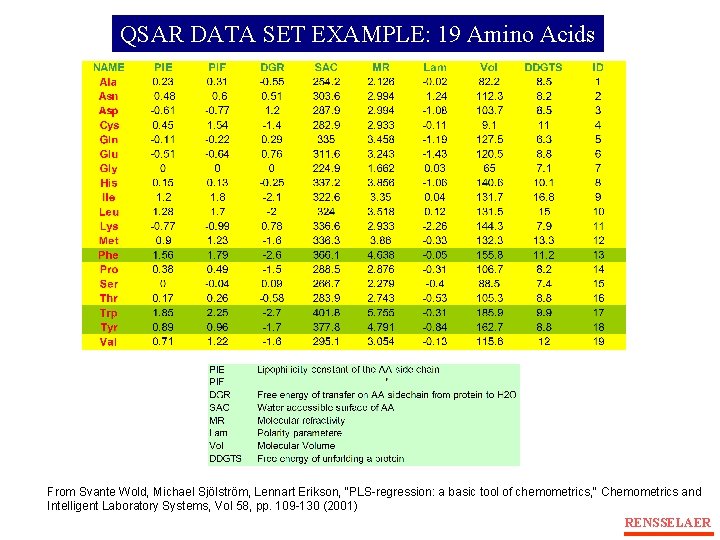
QSAR DATA SET EXAMPLE: 19 Amino Acids From Svante Wold, Michael Sjölström, Lennart Erikson, "PLS-regression: a basic tool of chemometrics, " Chemometrics and Intelligent Laboratory Systems, Vol 58, pp. 109 -130 (2001) RENSSELAER


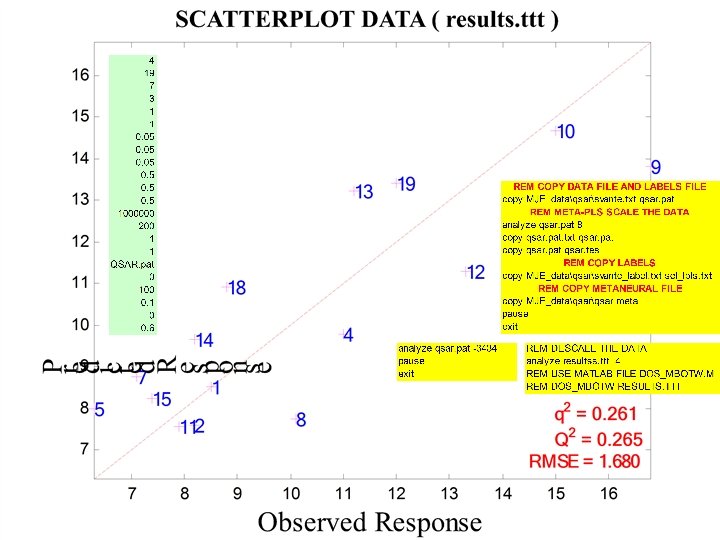
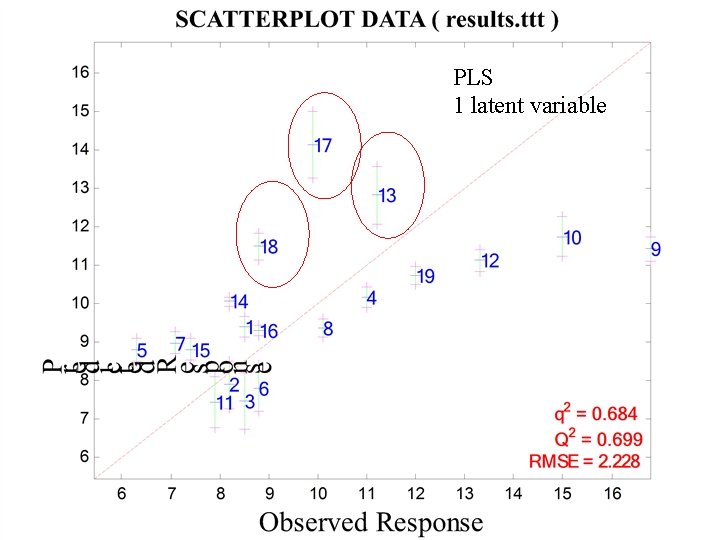
PLS 1 latent variable

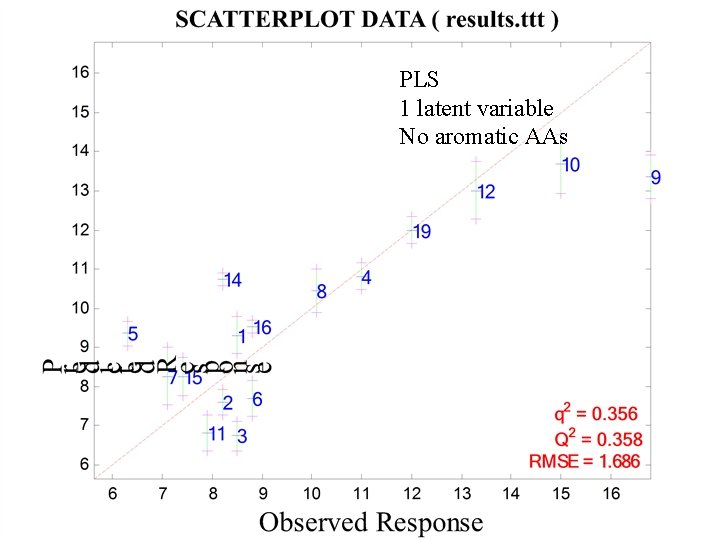
PLS 1 latent variable No aromatic AAs

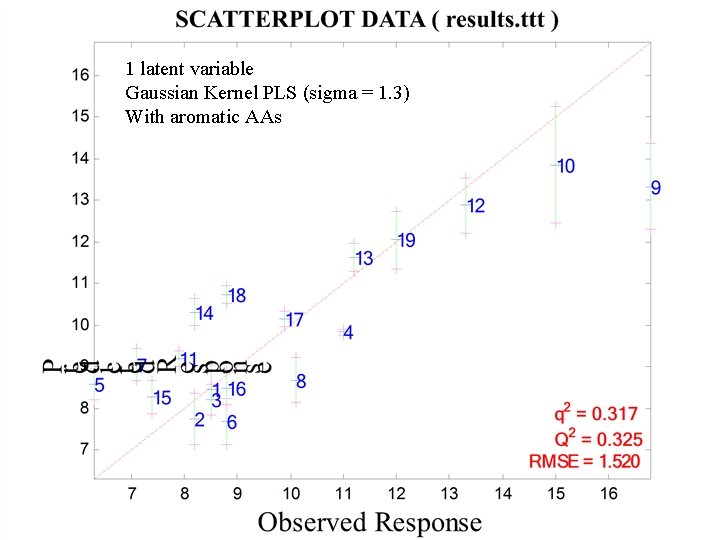
1 latent variable Gaussian Kernel PLS (sigma = 1. 3) With aromatic AAs

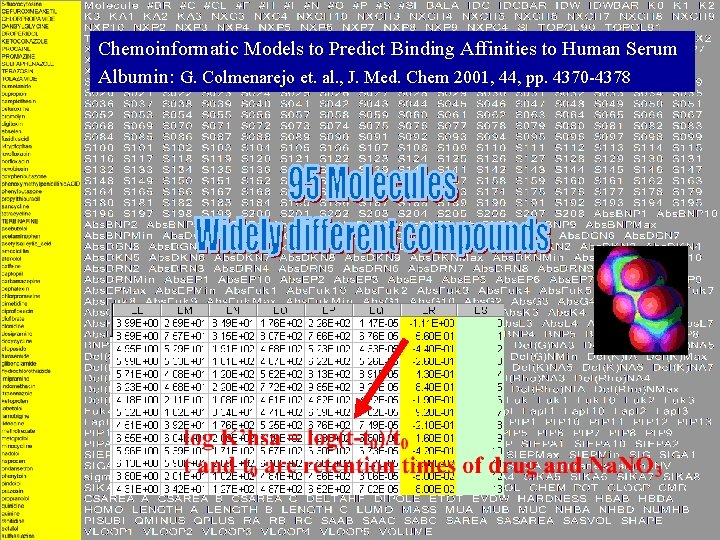
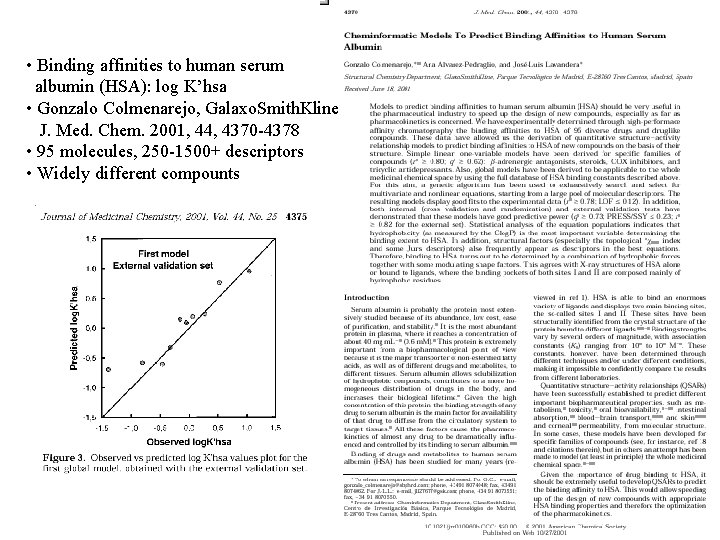
Chemoinformatic Models to Predict Binding Affinities to Human Serum Albumin: G. Colmenarejo et. al. , J. Med. Chem 2001, 44, pp. 4370 -4378

• Binding affinities to human serum albumin (HSA): log K’hsa • Gonzalo Colmenarejo, Galaxo. Smith. Kline J. Med. Chem. 2001, 44, 4370 -4378 • 95 molecules, 250 -1500+ descriptors • Widely different compounts

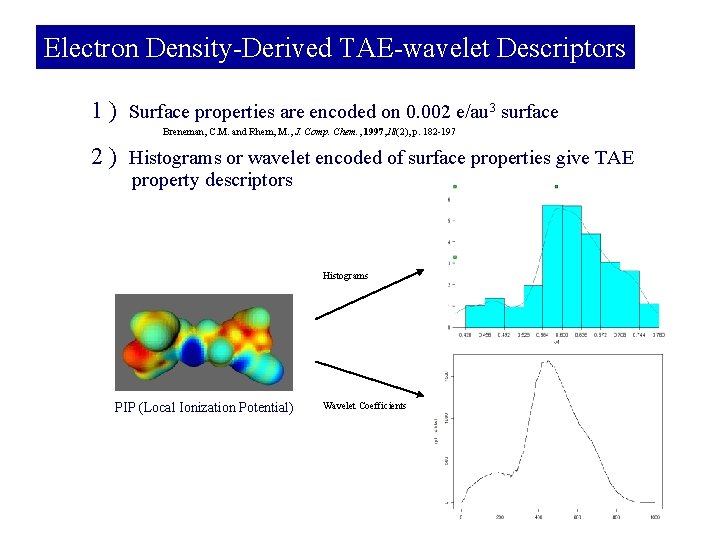
Electron Density-Derived TAE-wavelet Descriptors 1 ) Surface properties are encoded on 0. 002 e/au 3 surface Breneman, C. M. and Rhem, M. , J. Comp. Chem. , 1997, 18(2), p. 182 -197 2 ) Histograms or wavelet encoded of surface properties give TAE property descriptors Histograms PIP (Local Ionization Potential) Wavelet Coefficients

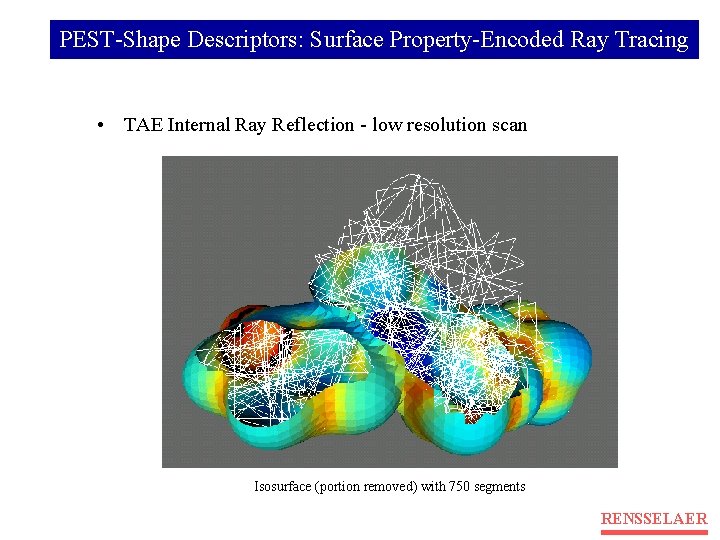
PEST-Shape Descriptors: Surface Property-Encoded Ray Tracing • TAE Internal Ray Reflection - low resolution scan Isosurface (portion removed) with 750 segments RENSSELAER

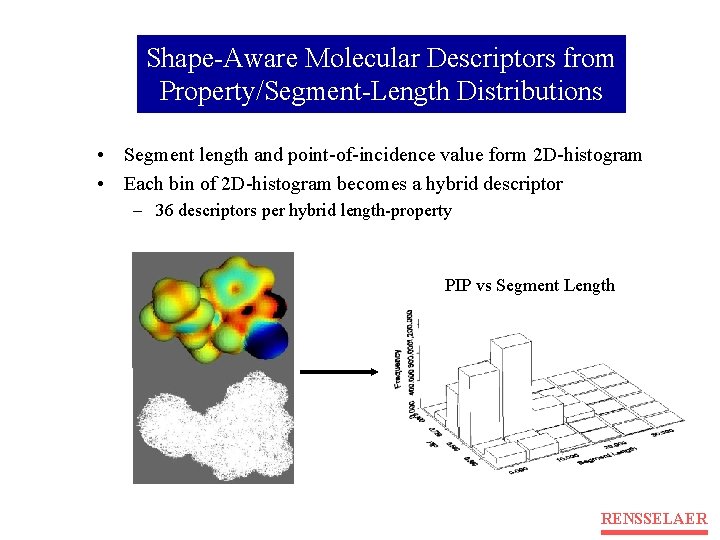
Shape-Aware Molecular Descriptors from Property/Segment-Length Distributions • Segment length and point-of-incidence value form 2 D-histogram • Each bin of 2 D-histogram becomes a hybrid descriptor – 36 descriptors per hybrid length-property PIP vs Segment Length RENSSELAER

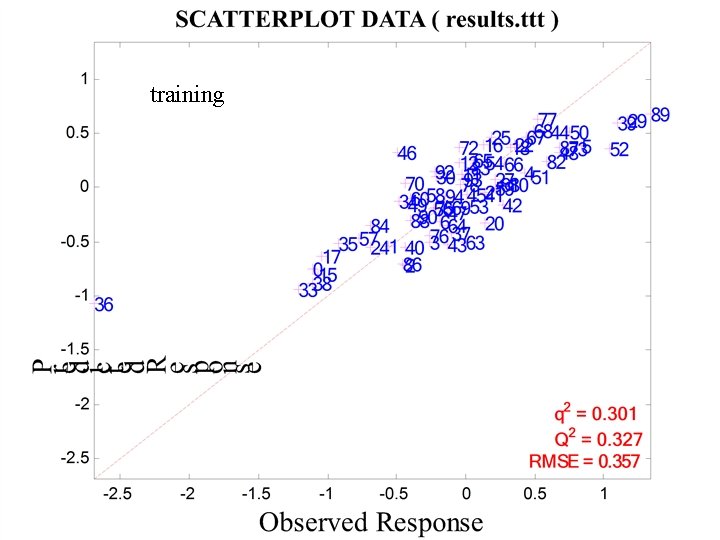
training

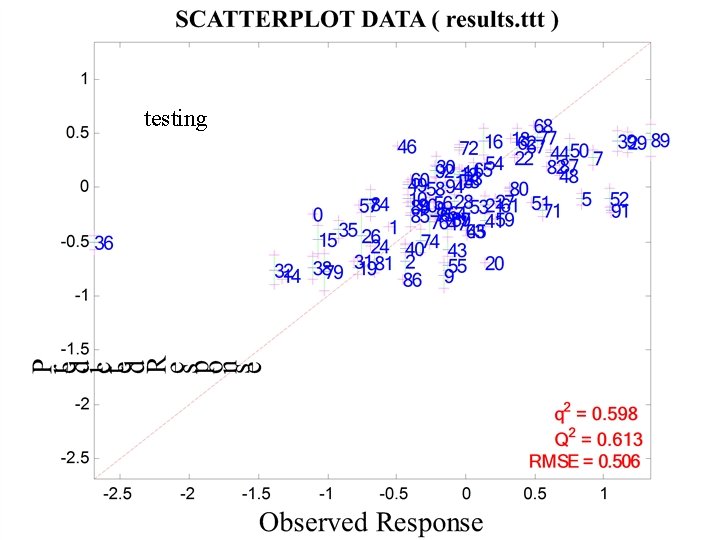
testing




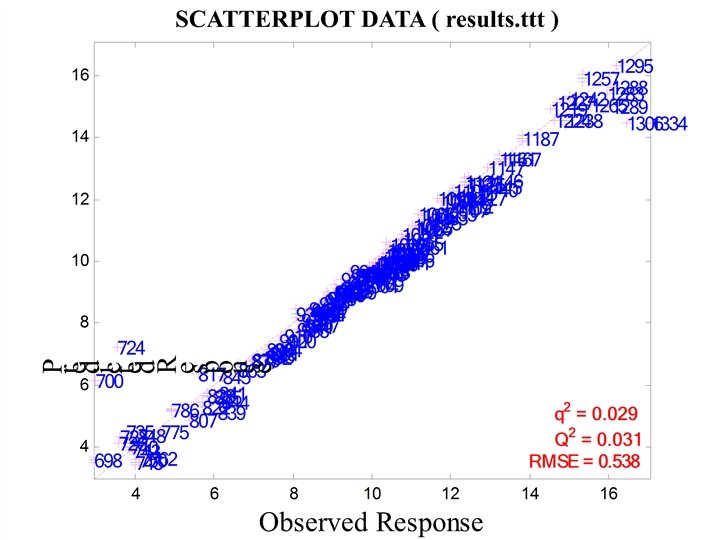
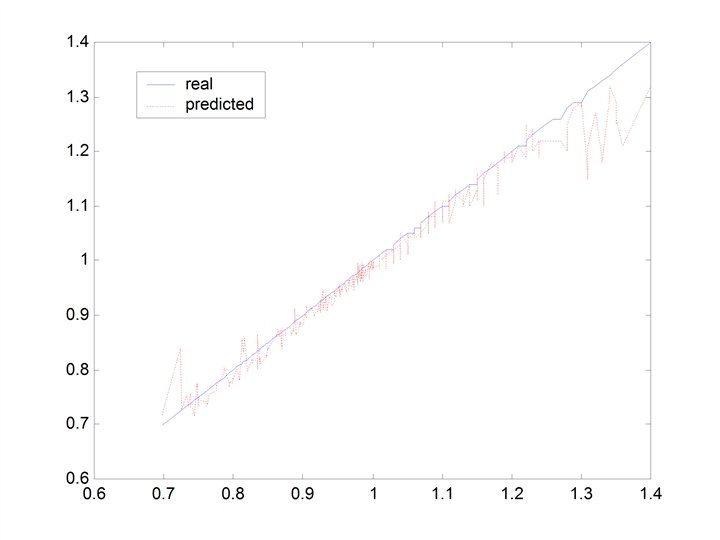
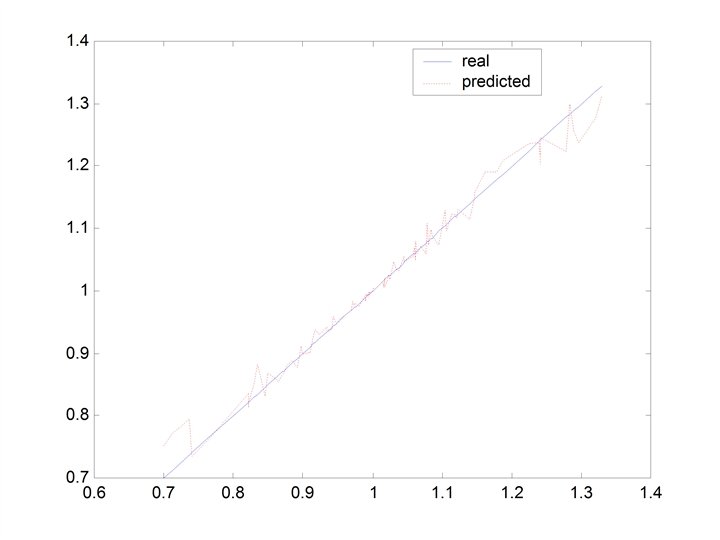
CHERKASSKY’S NONLINEAR BENCHMARK DATA • Generate 500 datapoints (400 training; 100 testing) for: Cherkas. bat




Y=sin|x|/|x| • Generate 500 datapoints (100 training; 500 testing) for:

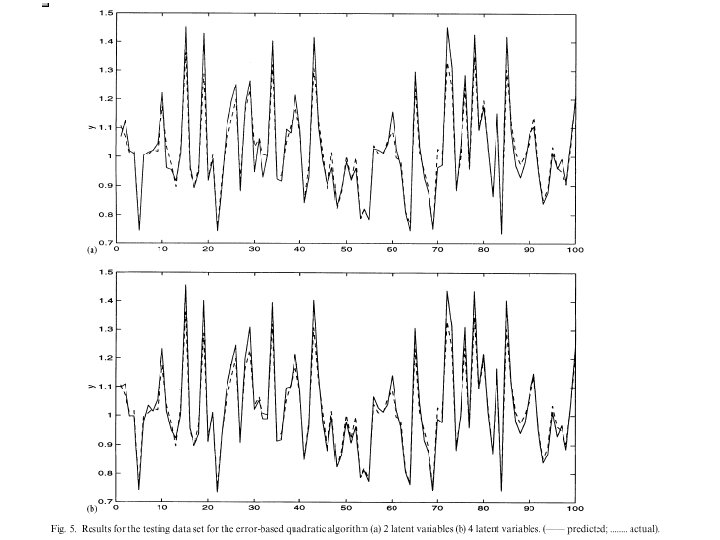
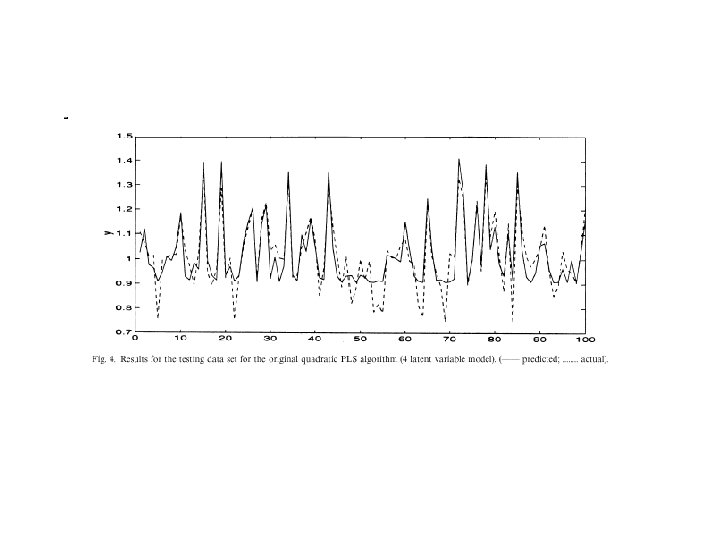
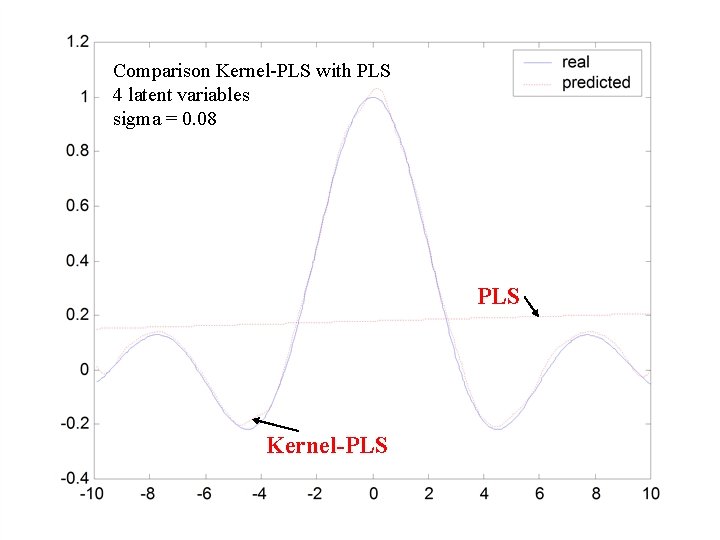
Comparison Kernel-PLS with PLS 4 latent variables sigma = 0. 08 PLS Kernel-PLS
