RNA Tertiary Structure Additional Motifs of Tertiary Structure




- Slides: 33

RNA Tertiary Structure

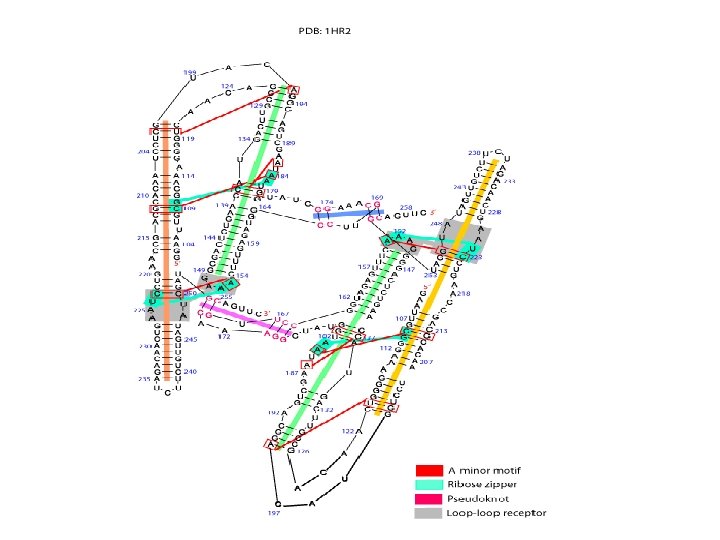
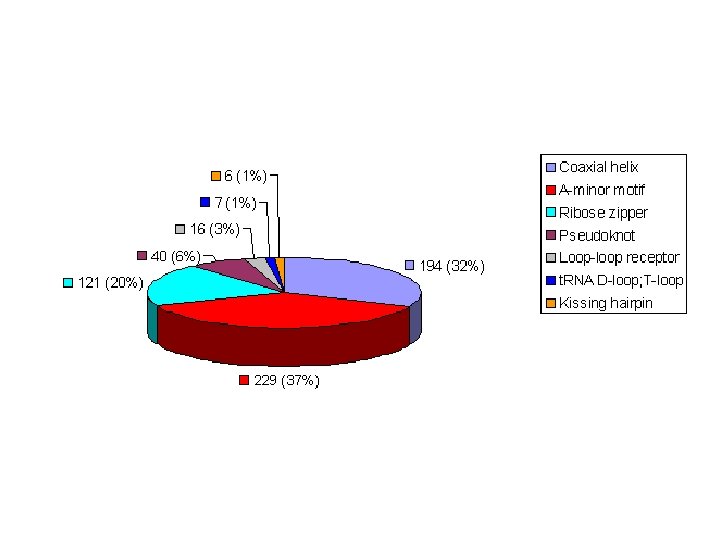
Additional Motifs of Tertiary Structure • • Coaxial helix A minor motif Pseudoknots Tetraloops Loop-loop Ribose zipper Kink turn motif

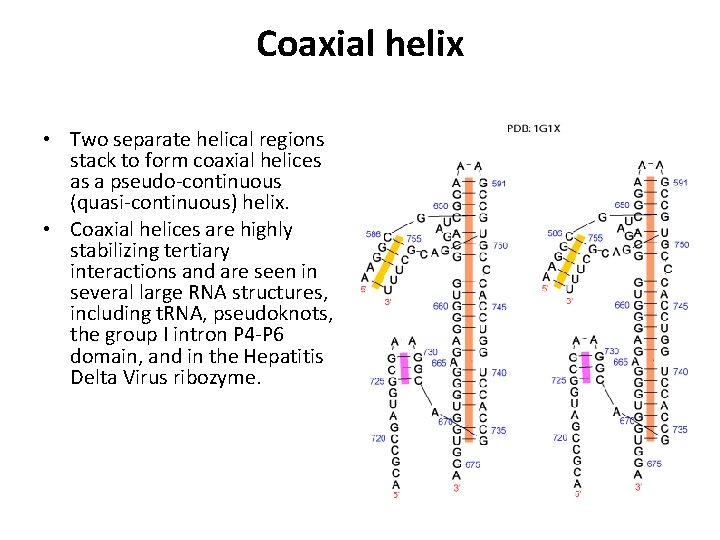
Coaxial helix • Two separate helical regions stack to form coaxial helices as a pseudo-continuous (quasi-continuous) helix. • Coaxial helices are highly stabilizing tertiary interactions and are seen in several large RNA structures, including t. RNA, pseudoknots, the group I intron P 4 -P 6 domain, and in the Hepatitis Delta Virus ribozyme.

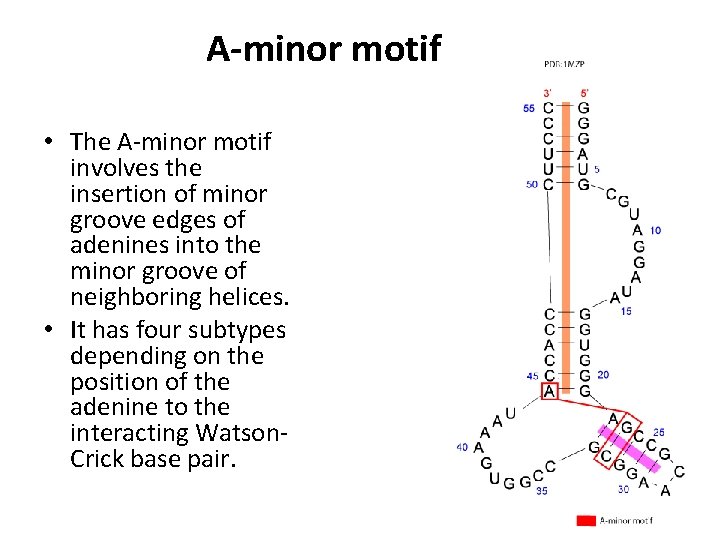
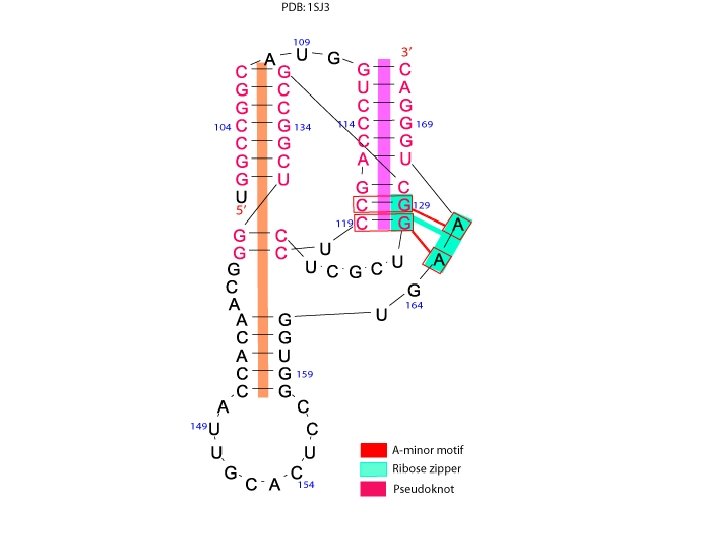
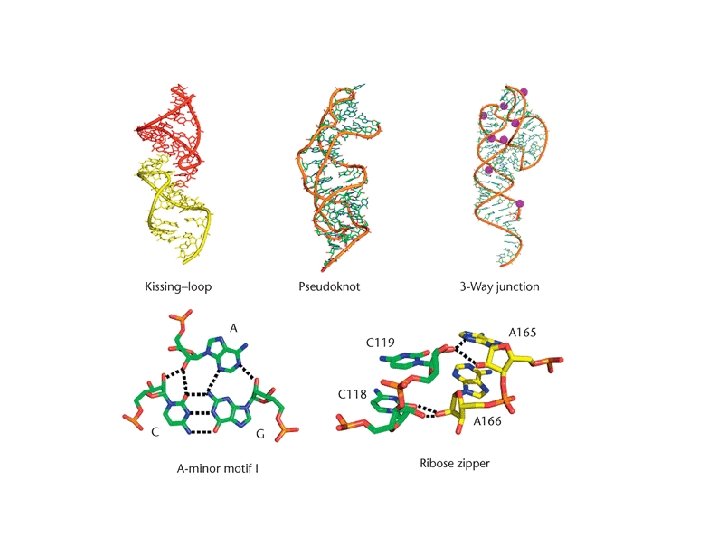
A-minor motif • The A-minor motif involves the insertion of minor groove edges of adenines into the minor groove of neighboring helices. • It has four subtypes depending on the position of the adenine to the interacting Watson. Crick base pair.

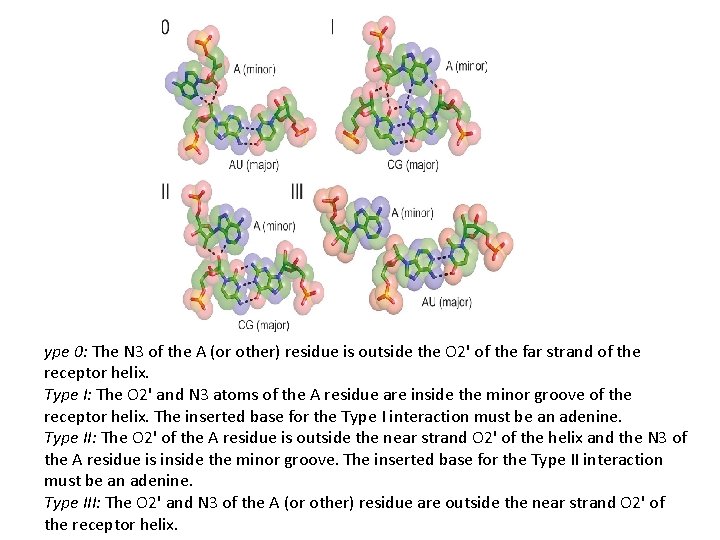
ype 0: The N 3 of the A (or other) residue is outside the O 2' of the far strand of the receptor helix. Type I: The O 2' and N 3 atoms of the A residue are inside the minor groove of the receptor helix. The inserted base for the Type I interaction must be an adenine. Type II: The O 2' of the A residue is outside the near strand O 2' of the helix and the N 3 of the A residue is inside the minor groove. The inserted base for the Type II interaction must be an adenine. Type III: The O 2' and N 3 of the A (or other) residue are outside the near strand O 2' of the receptor helix.

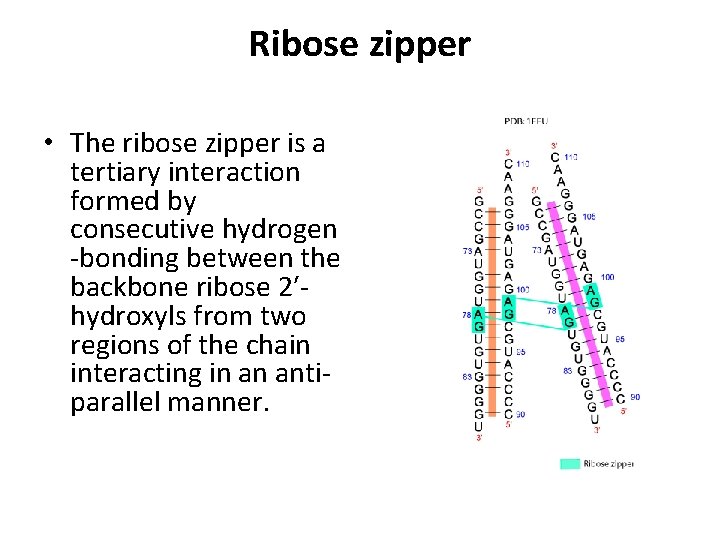
Ribose zipper • The ribose zipper is a tertiary interaction formed by consecutive hydrogen -bonding between the backbone ribose 2′hydroxyls from two regions of the chain interacting in an antiparallel manner.


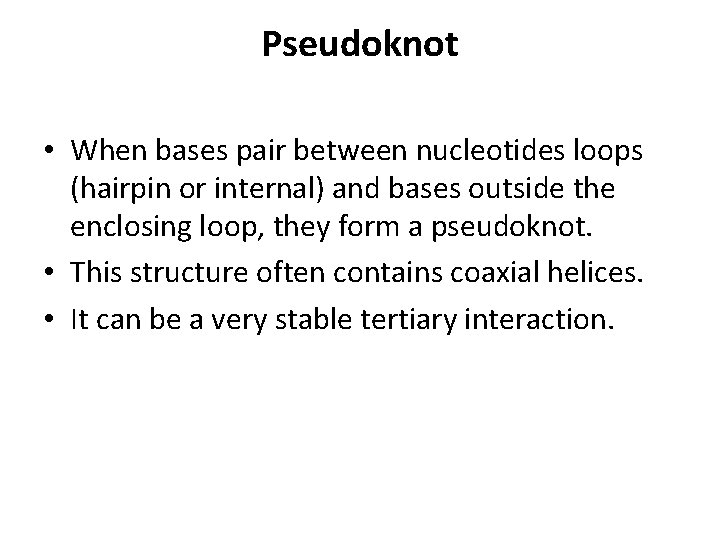
Pseudoknot • When bases pair between nucleotides loops (hairpin or internal) and bases outside the enclosing loop, they form a pseudoknot. • This structure often contains coaxial helices. • It can be a very stable tertiary interaction.


Loop-loop receptor • The tetraloop-tetraloop receptor was identified by comparative sequence analysis. • This tertiary interaction is characterized by specific hydrogen-bonding interactions between a tetraloop and a 11 -nucleotide internal loop/helical region that forms the receptor. • Other kinds of loop and receptor interactions, such as penta-loop/receptor and hexaloop/receptor, are observed so this motif is call loop-loop receptor.

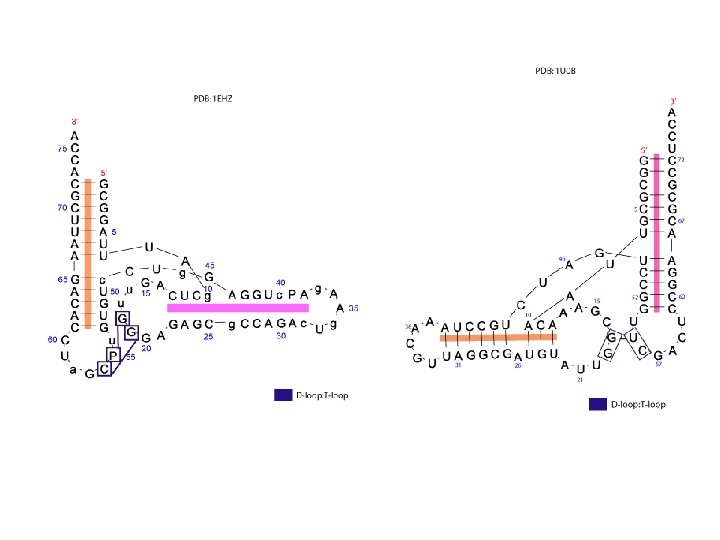
t. RNA D-loop; T-loop • The D-loop in t. RNA contains the modified nucleotide dihydrouridine. • It is composed of 7 to 11 bases and is closed by a Watson Crick base pair. The TψC-loop (generally called the T-loop) contains thymine, a base usually found in DNA and pseudouracil (ψ). • The D-loop and T-loop form a tertiary interaction in t. RNA.


Kissing hairpin • The kissing hairpin complex is a tertiary interaction formed by base pairing between the single-stranded residues of two hairpin loops with complementary sequences



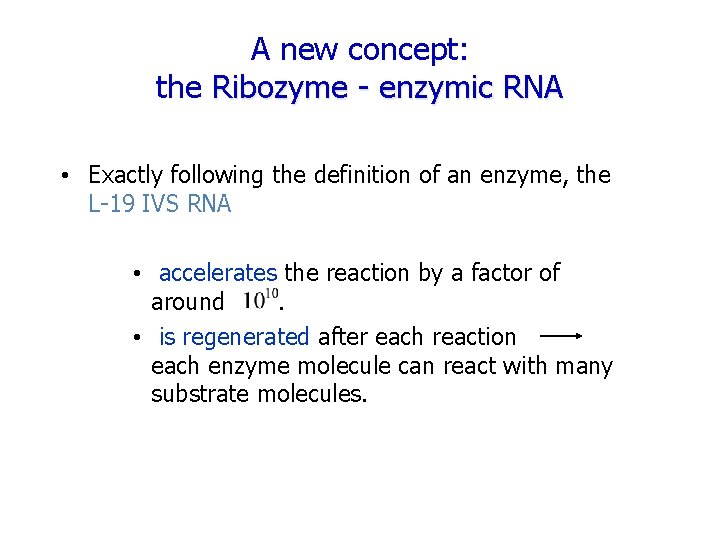
A new concept: the Ribozyme - enzymic RNA • Exactly following the definition of an enzyme, the L-19 IVS RNA • accelerates the reaction by a factor of around. • is regenerated after each reaction each enzyme molecule can react with many substrate molecules.

Ribozymes - Therapeutic Applications • Simple structure, site-specific cleavage activity and catalytic capability, make ribozymes effective modulators of gene expression. • Ribozyme-mediated gene modulation can target cancer cells, foreign genes that cause infectious diseases as well as other target sites (current research), and thereby alter the cellular pathology.

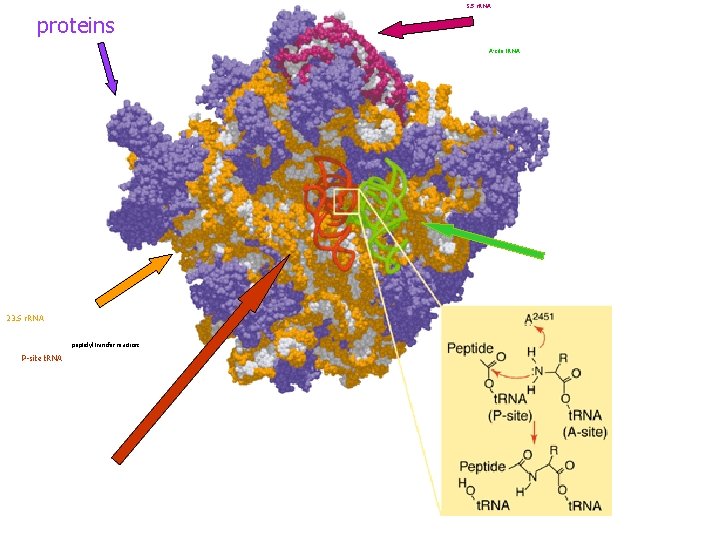
5 S r. RNA proteins A-site t. RNA 23 S r. RNA peptidyl transfer reaction: P-site t. RNA

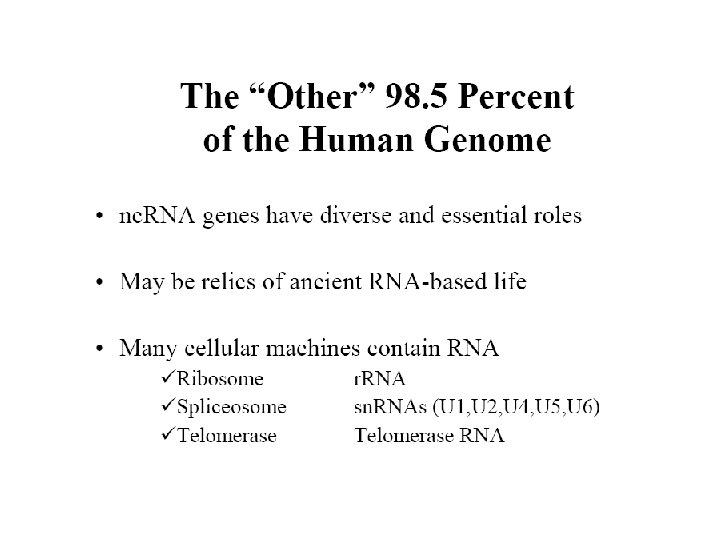
Ninety eight percent of the human genome does not code for protein. What is its function?


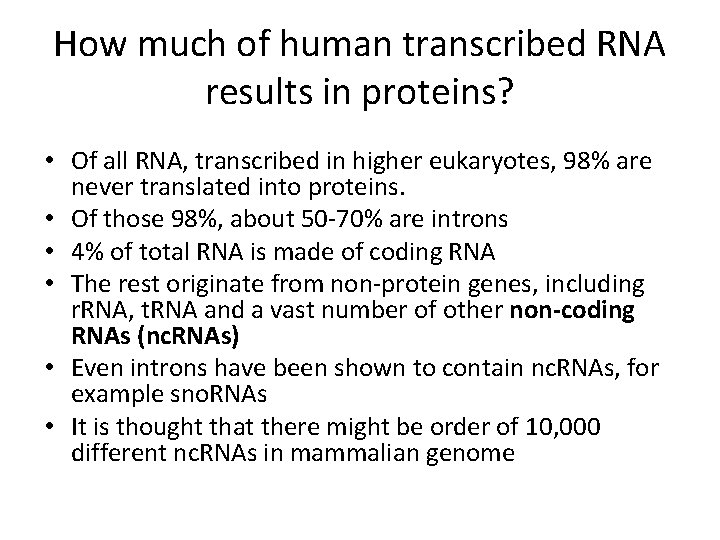
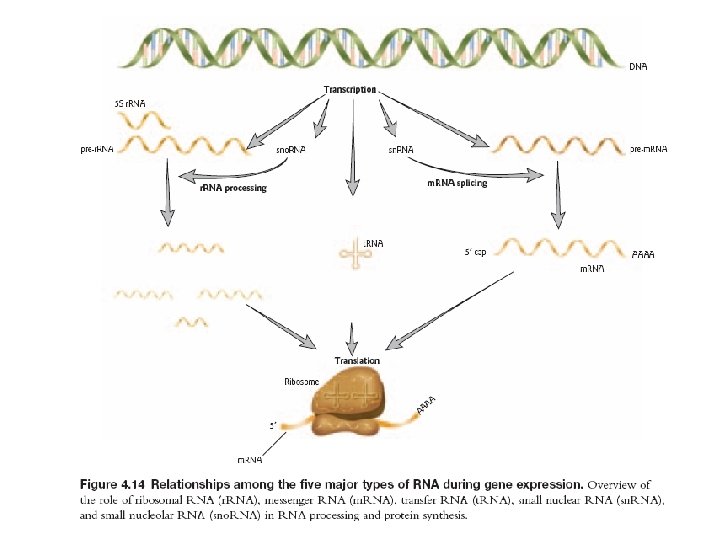
How much of human transcribed RNA results in proteins? • Of all RNA, transcribed in higher eukaryotes, 98% are never translated into proteins. • Of those 98%, about 50 -70% are introns • 4% of total RNA is made of coding RNA • The rest originate from non-protein genes, including r. RNA, t. RNA and a vast number of other non-coding RNAs (nc. RNAs) • Even introns have been shown to contain nc. RNAs, for example sno. RNAs • It is thought that there might be order of 10, 000 different nc. RNAs in mammalian genome

RNA functions • Storage/transfer of genetic information • Structural • Catalytic • Regulatory 22

RNA functions Storage/transfer of genetic information • Genomes • many viruses have RNA genomes single-stranded (ss. RNA) e. g. , retroviruses (HIV) double-stranded (ds. RNA) • Transfer of genetic information • m. RNA = "coding RNA" - encodes proteins D Dobbs ISU - BCB 444/544 X: RNA Structure & Function 23

RNA functions Structural • e. g. , r. RNA, which is major structural component of ribosomes BUT - its role is not just structural, also: Catalytic RNA in ribosome has peptidyltransferase activity • Enzymatic activity responsible for peptide bond formation between amino acids in growing peptide chain • Also, many small RNAs are enzymes "ribozymes” D Dobbs ISU - BCB 444/544 X: RNA Structure & Function 24

RNA functions Regulatory Recently discovered important new roles for RNAs In normal cells: • in "defense" - esp. in plants • in normal development e. g. , si. RNAs, mi. RNA As tools: • for gene therapy or to modify gene expression • RNA aptamers 25

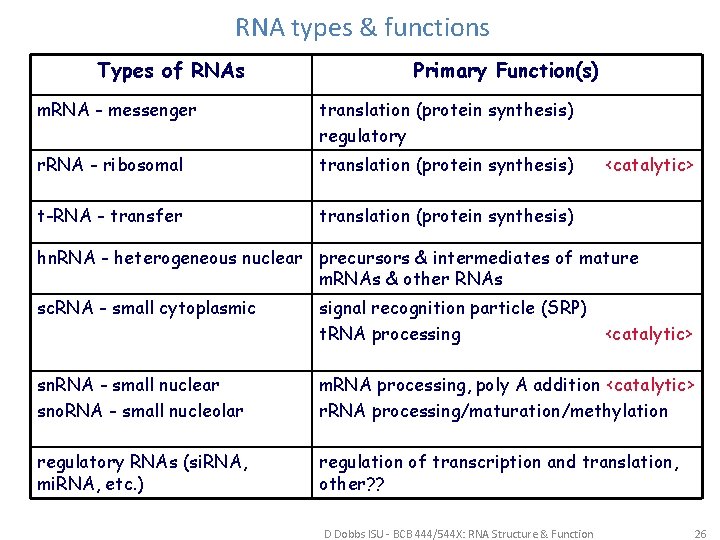
RNA types & functions Types of RNAs Primary Function(s) m. RNA - messenger translation (protein synthesis) regulatory r. RNA - ribosomal translation (protein synthesis) t-RNA - transfer translation (protein synthesis) <catalytic> hn. RNA - heterogeneous nuclear precursors & intermediates of mature m. RNAs & other RNAs sc. RNA - small cytoplasmic signal recognition particle (SRP) t. RNA processing <catalytic> sn. RNA - small nuclear sno. RNA - small nucleolar m. RNA processing, poly A addition <catalytic> r. RNA processing/maturation/methylation regulatory RNAs (si. RNA, mi. RNA, etc. ) regulation of transcription and translation, other? ? D Dobbs ISU - BCB 444/544 X: RNA Structure & Function 26

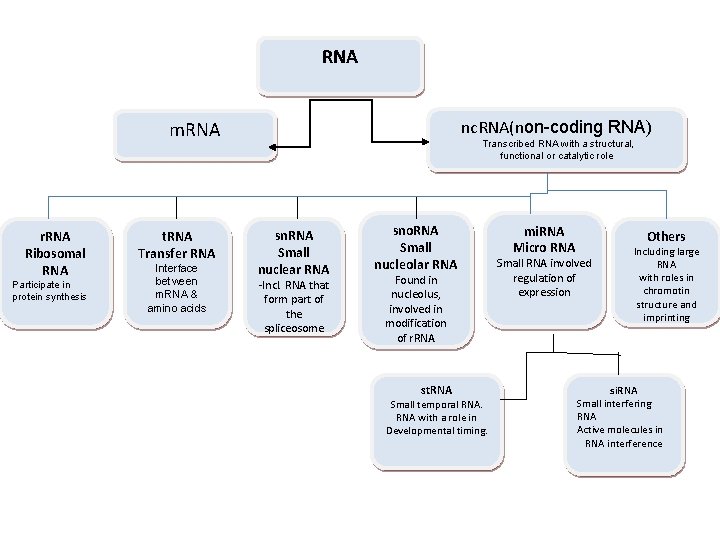
RNA nc. RNA(non-coding RNA) m. RNA r. RNA Ribosomal RNA Participate in protein synthesis t. RNA Transfer RNA Interface between m. RNA & amino acids Transcribed RNA with a structural, functional or catalytic role sn. RNA Small nuclear RNA -Incl. RNA that form part of the spliceosome sno. RNA Small nucleolar RNA Found in nucleolus, involved in modification of r. RNA st. RNA Small temporal RNA with a role in Developmental timing. mi. RNA Micro RNA Others Small RNA involved regulation of expression Including large RNA with roles in chromotin structure and imprinting si. RNA Small interfering RNA Active molecules in RNA interference


Small Nuclear RNAs • One important subcategory of small regulatory RNAs consists of the molecules know n as small nuclear RNAs (sn. RNAs). • These molecules play a critical role in gene regulation by w ay of RNA splicing. • sn. RNAs are found in the nucleus and are typically tightly bound to proteins in complexes called sn. RNPs (small nuclear ribonucleoproteins, sometimes pronounced "snurps"). • The most abundant of these molecules are the U 1, U 2, U 5, and U 4/U 6 particles, w hich are involved in splicing pre-m. RNA to give rise to mature m. RNA

Micro. RNAs • RNAs that are approximately 22 to 26 nucleotides in length. • The existence of mi. RNAs and their functions in gene regulation w ere initially discovered in the nematode C. Elegans. • Have also been found in many other species, including flies, mice, and humans. Several hundred mi. RNAs have been identified thus far, and many more may exist. • mi. RNAs have been show n to inhibit gene expression by repressing translation. • For example, the mi. RNAs encoded by C. elegans, lin-4 and let-7, bind to the 3' untranslated region of their target m. RNAs, preventing functional proteins from being produced during certain stages of larval development. • Additional studies indicate that mi. RNAs also play significant roles in cancer and other diseases. For example, the species mi. R-155 is enriched in B cells derived from Burkitt's lymphoma, and its sequence also correlates w ith a know n chromosomal translocation (exchange of DNA between chromosomes).

Small Interfering RNAs • Although these molecules are only 21 to 25 base pairs in length, they also work to inhibit gene expression. • si. RNAs were first defined by their participation in RNA interference (RNAi). They may have evolved as a defense mechanism against double-stranded RNA viruses. • si. RNAs are derived from longer transcripts in a process similar to that by which mi. RNAs are derived, and processing of both types of RNA involves the same enzyme, Dicer. • The two classes appear to be distinguished by their mechanisms of repression, but exceptions have been found in which si. RNAs exhibit behavior more typical of mi. RNAs, and vice versa.

mi. RNA Challenges for Computational Biology • Find the genes encoding micro. RNAs • Predict their regulatory targets Computational Prediction of Micro. RNA Genes & Targets • Integrate mi. RNAs into gene regulatory pathways & networks Need to modify traditional paradigm of "transcriptional control" by protein -DNA interactions to include mi. RNA regulatory mechanisms C Burge 2005 D Dobbs ISU - BCB 444/544 X: RNA Structure & Function 32

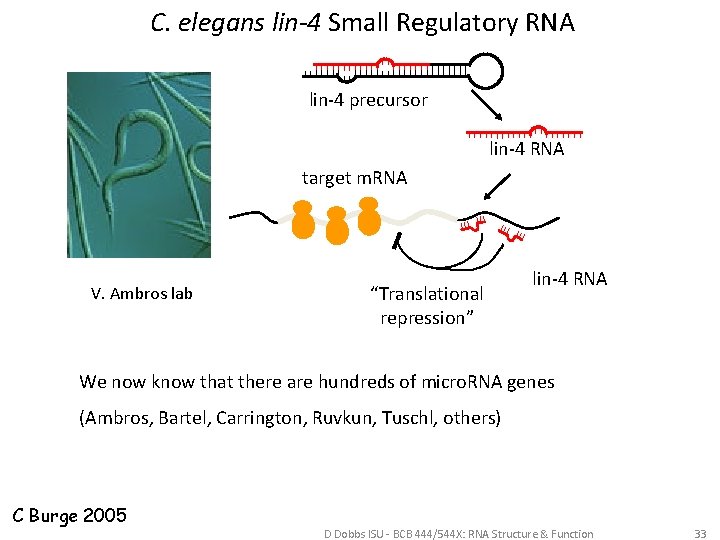
C. elegans lin-4 Small Regulatory RNA lin-4 precursor lin-4 RNA target m. RNA V. Ambros lab “Translational repression” lin-4 RNA We now know that there are hundreds of micro. RNA genes (Ambros, Bartel, Carrington, Ruvkun, Tuschl, others) C Burge 2005 D Dobbs ISU - BCB 444/544 X: RNA Structure & Function 33