RNA Seq data analysis using Variant Pipeline Tools





![ini style spec file format # Copyright. . . [pipeline description] description= human_hg 19_description= ini style spec file format # Copyright. . . [pipeline description] description= human_hg 19_description=](https://slidetodoc.com/presentation_image_h/372ee2c5b3c5801c327bdafecab38c34/image-25.jpg)
![Command line arguments # Copyright. . . [DEFAULT] java=java_comment=Full path to java if java Command line arguments # Copyright. . . [DEFAULT] java=java_comment=Full path to java if java](https://slidetodoc.com/presentation_image_h/372ee2c5b3c5801c327bdafecab38c34/image-26.jpg)
![Pipeline Variables [pipeline description] IGENOME_RESOURCE_DIR=${LOCAL_RESOURCE}/ pipeline_resource/i. Genome IGENOME_URL=ftp: //igenome: G 3 nom 3 s Pipeline Variables [pipeline description] IGENOME_RESOURCE_DIR=${LOCAL_RESOURCE}/ pipeline_resource/i. Genome IGENOME_URL=ftp: //igenome: G 3 nom 3 s](https://slidetodoc.com/presentation_image_h/372ee2c5b3c5801c327bdafecab38c34/image-27.jpg)
![Lambda function of variables [human_hg 19_1] action=Terminate. If(cond=${OUTPUT_DIR: not OUTPUT_DIR or os. path. isfile(OUTPUT_DIR)}, Lambda function of variables [human_hg 19_1] action=Terminate. If(cond=${OUTPUT_DIR: not OUTPUT_DIR or os. path. isfile(OUTPUT_DIR)},](https://slidetodoc.com/presentation_image_h/372ee2c5b3c5801c327bdafecab38c34/image-28.jpg)
![Input and input emitters [human_hg 19_201] input=${CMD_INPUT} input_emitter=Skip. If(select=${: %(sampling)s == 0}) Action= … Input and input emitters [human_hg 19_201] input=${CMD_INPUT} input_emitter=Skip. If(select=${: %(sampling)s == 0}) Action= …](https://slidetodoc.com/presentation_image_h/372ee2c5b3c5801c327bdafecab38c34/image-29.jpg)
![The Run. Command Action [align_400] input=${OUTPUT 301} action=Run. Command(cmd="%(java)s %(opt_java)s -jar %(picard_path)s/Merge. Sam. Files. The Run. Command Action [align_400] input=${OUTPUT 301} action=Run. Command(cmd="%(java)s %(opt_java)s -jar %(picard_path)s/Merge. Sam. Files.](https://slidetodoc.com/presentation_image_h/372ee2c5b3c5801c327bdafecab38c34/image-30.jpg)

- Slides: 33

RNA Seq data analysis using Variant Pipeline Tools Bo Peng, Ph. D. Department of Bioinformatics and Computational Biology The University of Texas, MD Anderson Cancer Center Jan 12, 2015

Why yet another pipeline tool?

Challenges for Bioinformatics Pipelines Annotation Availability and variability of resources (reference genomes, annotations) High demand in CPU, I/O, and/or memory usage, leading to high failure rate on cluster Difference in file formats and incompatibility with files in the same format Storage Pipeline Data • • Multiple frequently updated tools Inconsistency between different tools, even different versions of the same tools • • Large input, output and intermediate files Difference in file formats

Pipeline Implementations Script based Specification based • Shell, Perl, Python, … • Galaxy, Seq. Ware, Nimbix, … Pros: • Extremely flexible • Familiar to most researchers Pros: • Separation of logic and execution • Extremely powerful on cluster or cloud • Documentation is usually provided Cons: • Documentation is usually lacking • Usually lengthy • Difficult to read and modify • Less scalable • Usually lack of advanced execution control features Cons: • Platform/vendor-dependent • Can be difficult to install and execute • Steeper learning curve • XML-based configuration files can be difficult to write and modify • Some are not flexible and extensible

Variant Tools is a software tools for the integrated analysis of genetic variants for next-generation sequencing studies. Utilities Variant Pipeline Tools Variant Association Tools Variant Simulation Tools Variant Tools manipulation, annotation, and selection of variants Variant Association Tools rare variant association analysis Variant Simulation Tools Simulation of realistic samples with rare variants Variant Pipeline Tools Execution of variant tools and other bioinformatics pipelines. Utilities and reporting tools San Lucas et al. 2012, Wang et al. 2014, Peng 2014

Variant Pipeline Tools is a module of Variant Tools that provides a light-weight pipeline specification and execution mechanism for bioinformatics projects. VPT is designed to be: • Easy to use • Easy to share • Easy to read/write • Flexible • Extensible • Fault-tolerant

Features

Three commands to remember • vtools show pipelines • vtools show pipeline SPECFILE • vtools execute SPECFILE [PIPELINE] [options] • It can be time consuming to set up the running environment (install tools) and download resources • Sharing of environment and resources is possible but is administratively challenging

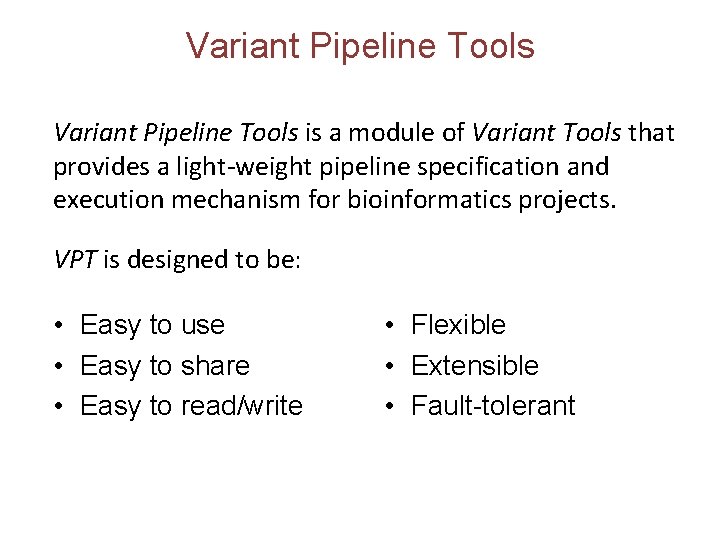
Show all available pipelines List all available spec files in the variant tools repositories

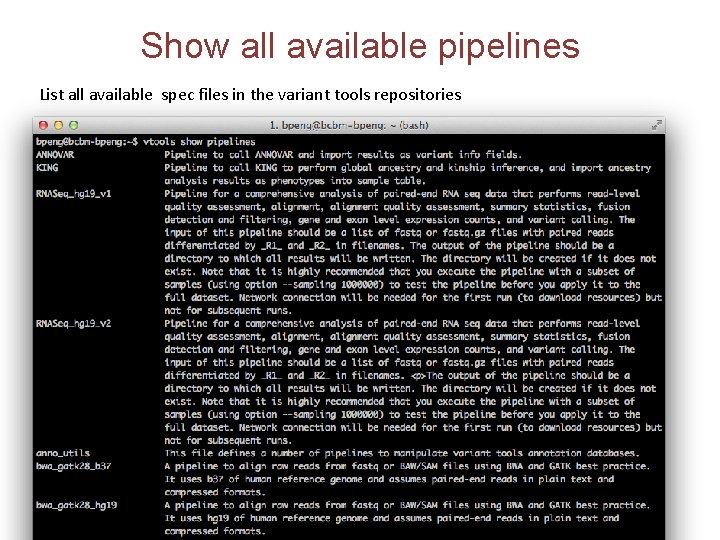
Show details of a local or online pipelines • Pipelines can be local or online • Online pipelines are automatically downloaded to users’ resource directory ($HOME/. variant_tools)

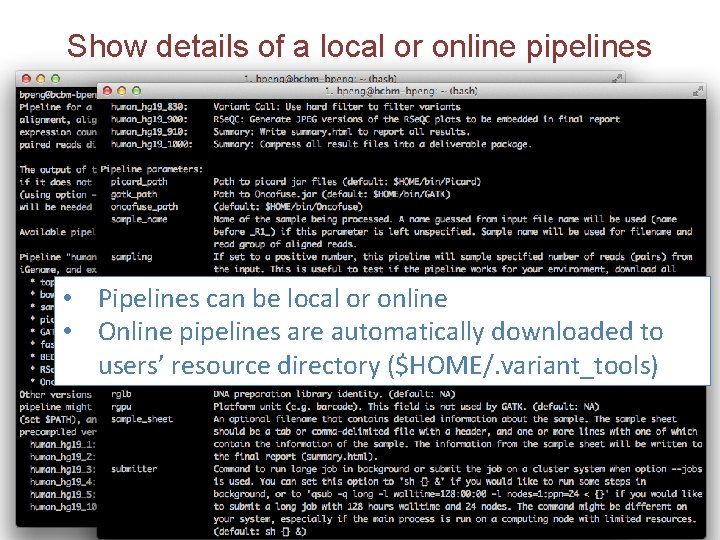
Execute a Pipeline

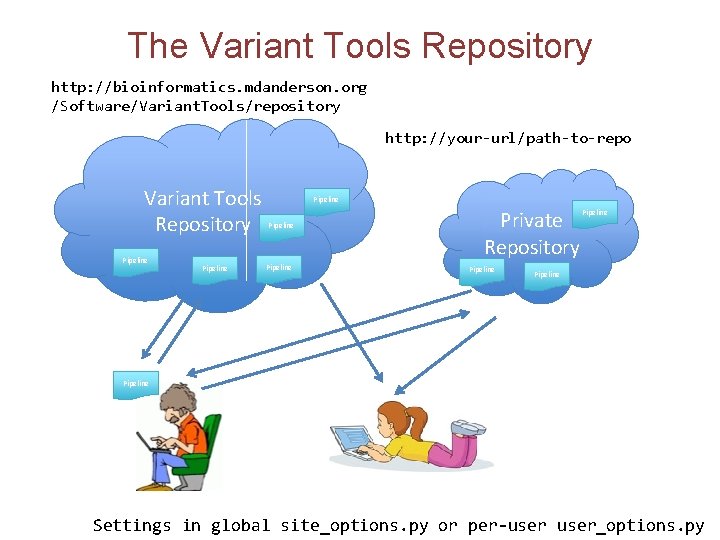
The Variant Tools Repository http: //bioinformatics. mdanderson. org /Software/Variant. Tools/repository http: //your-url/path-to-repo Variant Tools Repository Pipeline Pipeline Private Repository Pipeline Settings in global site_options. py or per-user_options. py

A Galaxy pipeline file

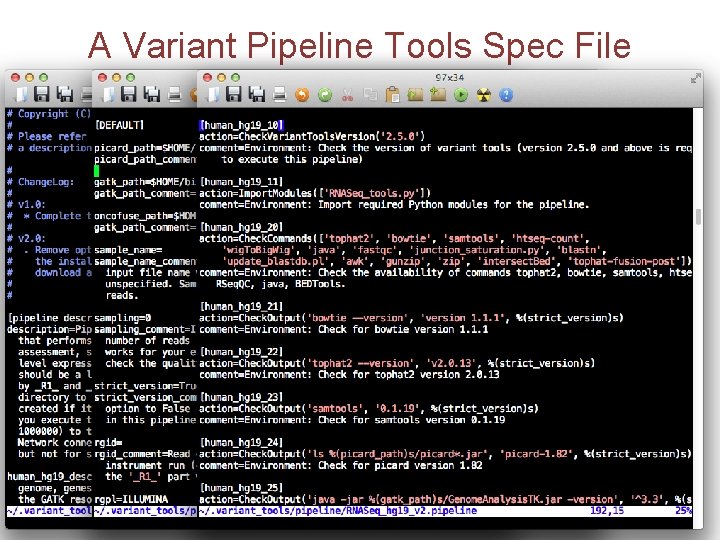
A Variant Pipeline Tools Spec File

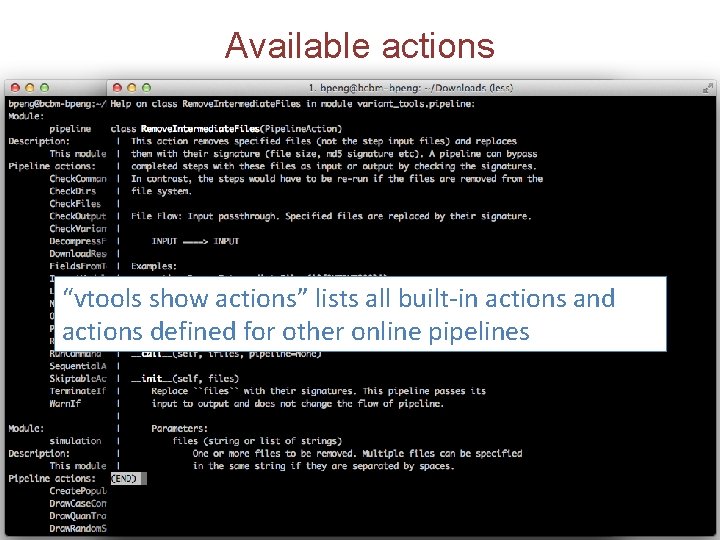
Allowed Actions • Check user input • Check availability and versions of commands • Check or download required resources • Execute arbitrary shell commands • Execute variant tools-provided, third-party, or self-written actions (Python based)

Available actions “vtools show actions” lists all built-in actions and actions defined for other online pipelines

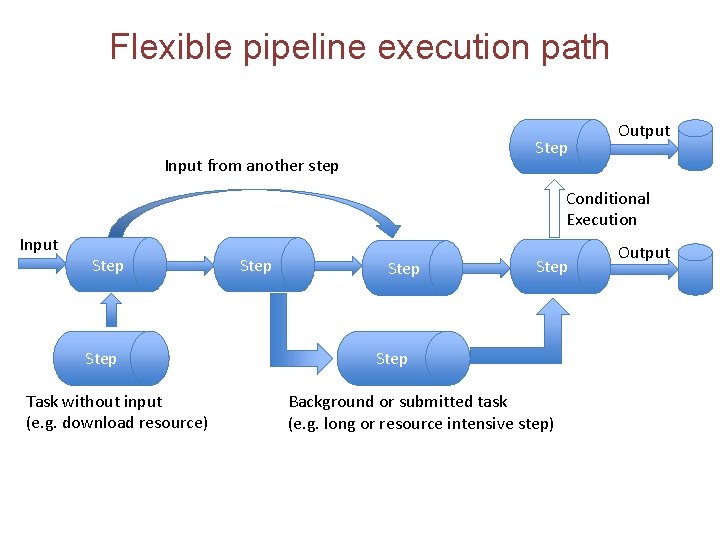
Flexible pipeline execution path Step Input from another step Output Conditional Execution Input Step Task without input (e. g. download resource) Step Background or submitted task (e. g. long or resource intensive step) Output

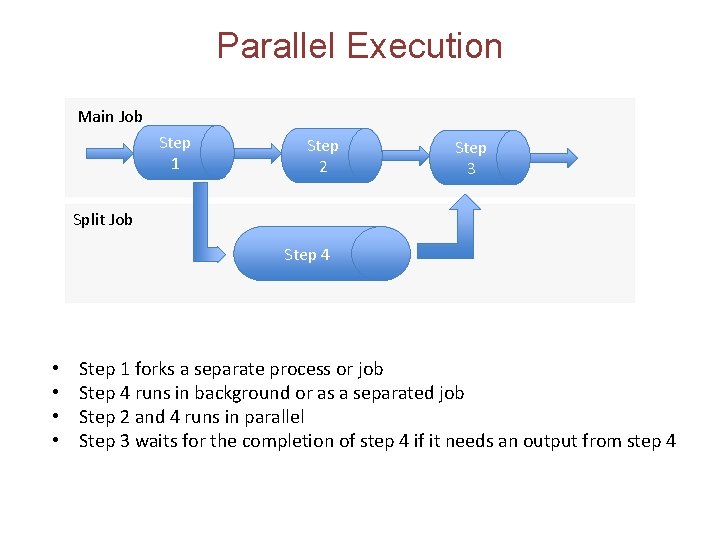
Parallel Execution Main Job Step 1 Step 2 Step 3 Split Job Step 4 • • Step 1 forks a separate process or job Step 4 runs in background or as a separated job Step 2 and 4 runs in parallel Step 3 waits for the completion of step 4 if it needs an output from step 4

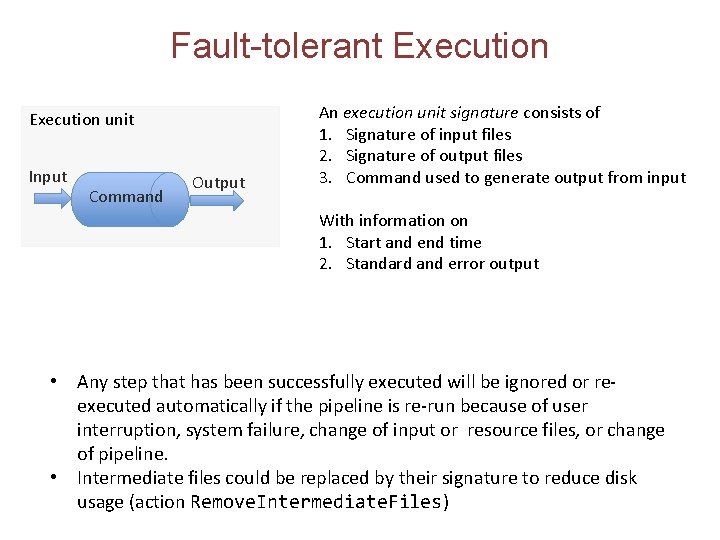
Fault-tolerant Execution unit Input Command Output An execution unit signature consists of 1. Signature of input files 2. Signature of output files 3. Command used to generate output from input With information on 1. Start and end time 2. Standard and error output • Any step that has been successfully executed will be ignored or reexecuted automatically if the pipeline is re-run because of user interruption, system failure, change of input or resource files, or change of pipeline. • Intermediate files could be replaced by their signature to reduce disk usage (action Remove. Intermediate. Files)

A comprehensive RNA Seq analysis pipeline

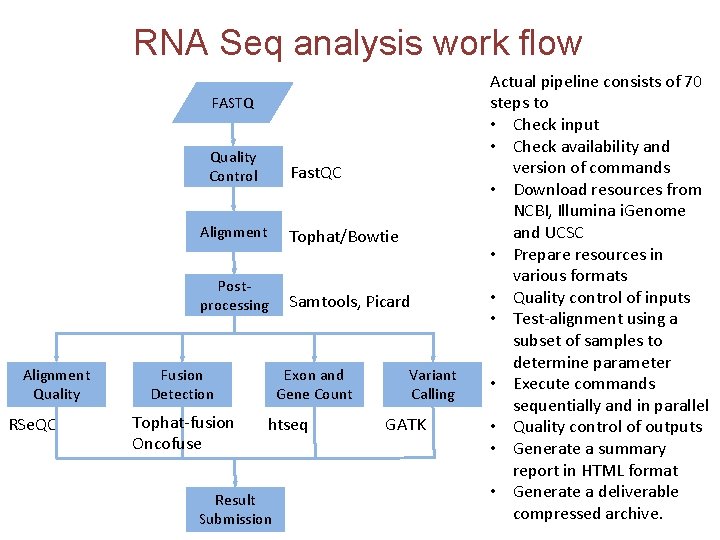
RNA Seq analysis work flow FASTQ Quality Control Alignment Quality RSe. QC Fast. QC Alignment Tophat/Bowtie Postprocessing Samtools, Picard Fusion Detection Tophat-fusion Oncofuse Exon and Gene Count htseq Result Submission Variant Calling GATK Actual pipeline consists of 70 steps to • Check input • Check availability and version of commands • Download resources from NCBI, Illumina i. Genome and UCSC • Prepare resources in various formats • Quality control of inputs • Test-alignment using a subset of samples to determine parameter • Execute commands sequentially and in parallel • Quality control of outputs • Generate a summary report in HTML format • Generate a deliverable compressed archive.

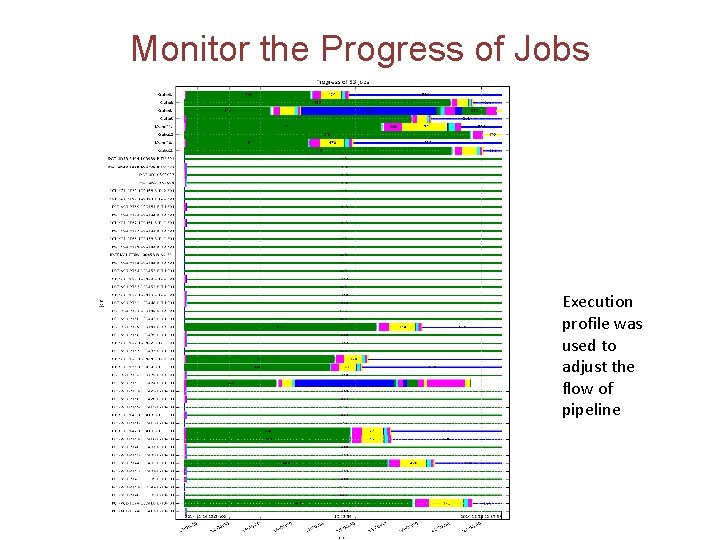
Monitor the Progress of Jobs Execution profile was used to adjust the flow of pipeline

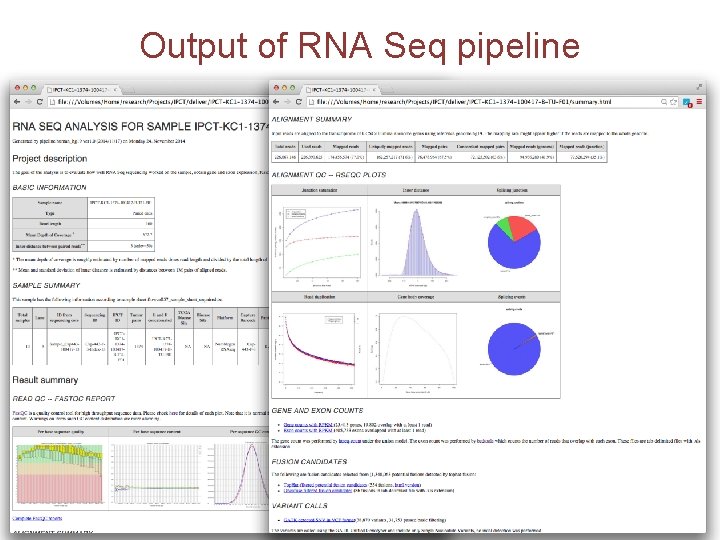
Output of RNA Seq pipeline

Technical Details (Please feel free to leave if you are not technically oriented)
![ini style spec file format Copyright pipeline description description humanhg 19description ini style spec file format # Copyright. . . [pipeline description] description= human_hg 19_description=](https://slidetodoc.com/presentation_image_h/372ee2c5b3c5801c327bdafecab38c34/image-25.jpg)
ini style spec file format # Copyright. . . [pipeline description] description= human_hg 19_description= Mouse_mm 10_description= [*_1] action=Check. Variant. Tools. Version(‘ 2. 5. 0’) comment=Environment: Check the version of variant tools [human_hg 19_10] action= comment= [human_hg 19_20] input= action= comment= [mouse_mm 10_10] Action= • One spec file can define multiple pipelines • Extended ini format • # as comment • Values can span multiple lines • Description and comment are part of the pipelines • Pipeline steps are defined in sections
![Command line arguments Copyright DEFAULT javajavacommentFull path to java if java Command line arguments # Copyright. . . [DEFAULT] java=java_comment=Full path to java if java](https://slidetodoc.com/presentation_image_h/372ee2c5b3c5801c327bdafecab38c34/image-26.jpg)
Command line arguments # Copyright. . . [DEFAULT] java=java_comment=Full path to java if java not in $PATH gatk_path= gatk_ath=Path to GATK (with Genome. Analysis. TK. jar) [human_hg 19_10] Action=Check. Output(‘%(java)s –version’, ‘ 1. 7. 0’) comment=Check version of java version 1. 7. 0 [human_hg 19_20] Action=Run. Command(‘%(java)s -jar %(gatk_path)s/Genome. Analysis. TK. jar …’) Comment=Execute GATK • Command line arguments are defined in section DEFAULT • Default values can be provided • Value %(NAME)s are replaced with default value or command line argument • % in other places should be written as %%
![Pipeline Variables pipeline description IGENOMERESOURCEDIRLOCALRESOURCE pipelineresourcei Genome IGENOMEURLftp igenome G 3 nom 3 s Pipeline Variables [pipeline description] IGENOME_RESOURCE_DIR=${LOCAL_RESOURCE}/ pipeline_resource/i. Genome IGENOME_URL=ftp: //igenome: G 3 nom 3 s](https://slidetodoc.com/presentation_image_h/372ee2c5b3c5801c327bdafecab38c34/image-27.jpg)
Pipeline Variables [pipeline description] IGENOME_RESOURCE_DIR=${LOCAL_RESOURCE}/ pipeline_resource/i. Genome IGENOME_URL=ftp: //igenome: G 3 nom 3 s 4 u@ussdftp. illumina. com/Homo_sapiens/UCSC/hg 19/ Homo_sapiens_UCSC_hg 19. tar. gz [human_hg 19_100] action=Download. Resource(resource='${IGENOME_URL}', dest_dir="${IGENOME_RESOURCE_DIR}") [human_hg 19_440] input=${INPUT 300} action=Run. Command('tophat 2 --zpacker 0 -no-coverage-search --num-threads 8 --GTF ${GENES_CHR_GTF} --segment-length 25. . . ’) • Pipeline variables keep runtime information of pipelines • Value %{NAME} are replaced with value of pipeline variable • Variables such as CMD_INPUT, CMD_OUTPUT are predefined • Variables such as INPUT 300 are set during the execution • Variables can be defined by pipeline actions
![Lambda function of variables humanhg 191 actionTerminate IfcondOUTPUTDIR not OUTPUTDIR or os path isfileOUTPUTDIR Lambda function of variables [human_hg 19_1] action=Terminate. If(cond=${OUTPUT_DIR: not OUTPUT_DIR or os. path. isfile(OUTPUT_DIR)},](https://slidetodoc.com/presentation_image_h/372ee2c5b3c5801c327bdafecab38c34/image-28.jpg)
Lambda function of variables [human_hg 19_1] action=Terminate. If(cond=${OUTPUT_DIR: not OUTPUT_DIR or os. path. isfile(OUTPUT_DIR)}, message='Please specify an output directory’) [human_hg 19_440] input=${INPUT 300} action=Run. Command('tophat 2 --segment-length 25 ${REFERENCE_DIR}/Bowtie. Index/genome ${INPUT: ', '. join(sorted([x for x in INPUT if '_R 1_' in x]))} ${INPUT: ', '. join(sorted([x for x in INPUT if '_R 2_' in x]))}', output='${ALIGNMENT_OUT}/accepted_hits. bam’) • Lambda function provides an extremely flexible way to use pipeline variables • Need basic understanding of Python • Lambda functions with 0, 1, or more parameters are acceptable
![Input and input emitters humanhg 19201 inputCMDINPUT inputemitterSkip Ifselect samplings 0 Action Input and input emitters [human_hg 19_201] input=${CMD_INPUT} input_emitter=Skip. If(select=${: %(sampling)s == 0}) Action= …](https://slidetodoc.com/presentation_image_h/372ee2c5b3c5801c327bdafecab38c34/image-29.jpg)
Input and input emitters [human_hg 19_201] input=${CMD_INPUT} input_emitter=Skip. If(select=${: %(sampling)s == 0}) Action= … comment=Draw a subset of samples if parameter --sampling is defined [align_200] input_emitter=Emit. Input(select=['bam', 'sam'], pass_unselected=False) comment=Convert bam files to paired fastq files if the input is in bam/sam format. [align_500] input_emitter=Emit. Input('single', select='fastq', pass_unselected=False) action=. . . • Input files could be passed to action altogether (default), one by one, in pairs, or ignored. • An emitter can be used to ignore a step, execute an alternative step if a previous step fails, or select files with matching types
![The Run Command Action align400 inputOUTPUT 301 actionRun Commandcmdjavas optjavas jar picardpathsMerge Sam Files The Run. Command Action [align_400] input=${OUTPUT 301} action=Run. Command(cmd="%(java)s %(opt_java)s -jar %(picard_path)s/Merge. Sam. Files.](https://slidetodoc.com/presentation_image_h/372ee2c5b3c5801c327bdafecab38c34/image-30.jpg)
The Run. Command Action [align_400] input=${OUTPUT 301} action=Run. Command(cmd="%(java)s %(opt_java)s -jar %(picard_path)s/Merge. Sam. Files. jar ${INPUT: ' '. join(['INPUT=' + x for x in INPUT])} USE_THREADING=true VALIDATION_STRINGENCY=LENIENT OUTPUT=${INPUT: INPUT[0][: -4] + '_merged. bam'}", output="${INPUT: INPUT[0][: -4] + '_merged. bam'}") [human_hg 19_300] action=Run. Command(['${INPUT: "gunzip -c" if INPUT[0]. endswith(". gz") else "cat"} ${INPUT} | fastqc stdin --outdir=${READQC_OUT}', 'mv ${READQC_OUT}/stdin_fastqc. html ${READQC_OUT}/${SAMPLENAME}_fastqc. html'], output='${READQC_OUT}/${SAMPLENAME}_fastqc. html', submitter='sh {} &') • Run one or more commands • Run in shell mode (IO pipes are allowed) • Record execution signature if output files are specified • Can be submitted to background

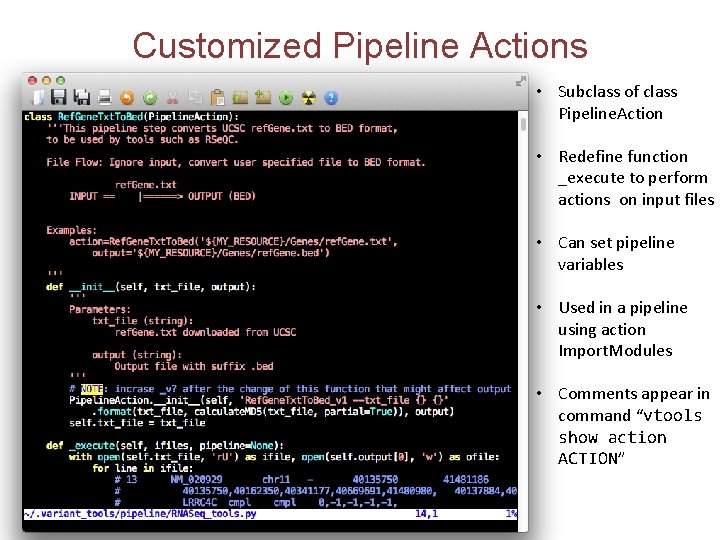
Customized Pipeline Actions • Subclass of class Pipeline. Action • Redefine function _execute to perform actions on input files • Can set pipeline variables • Used in a pipeline using action Import. Modules • Comments appear in command “vtools show action ACTION”

Summary Variant Pipeline Tools is designed to implement, share, and execute arbitrary (bioinformatics) pipelines. • It is (hopefully) easy to use, share, read/write, flexible, extensible, and fault-tolerant • It has been used by variant tools users for a while (but no one has shared their pipelines with us) However, • It is NOT designed to handle huge amount of data on the cloud (no features such as Map. Reduce) • It can be troublesome to maintain a large collection of pipelines

Acknowledgements • • Gao Wang Dr. Chris Amos Dr. Paul Scheet Dr. Biao Li Dr. Suzanne Leal Dr. John Weinstein and others • • • Grant 1 R 01 HG 005859 (Dr. Paul Scheet) The Prevent Cancer Foundation The Michael and Susan Dell Foundation The Chapman Foundation MD Anderson High Performance Computing Cluster