RNA Secondary Structure Prediction RNA role in organisms






- Slides: 48

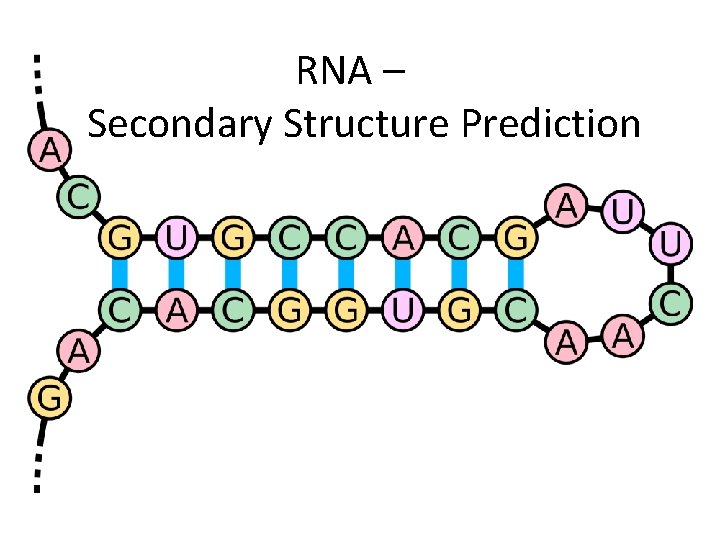
RNA – Secondary Structure Prediction

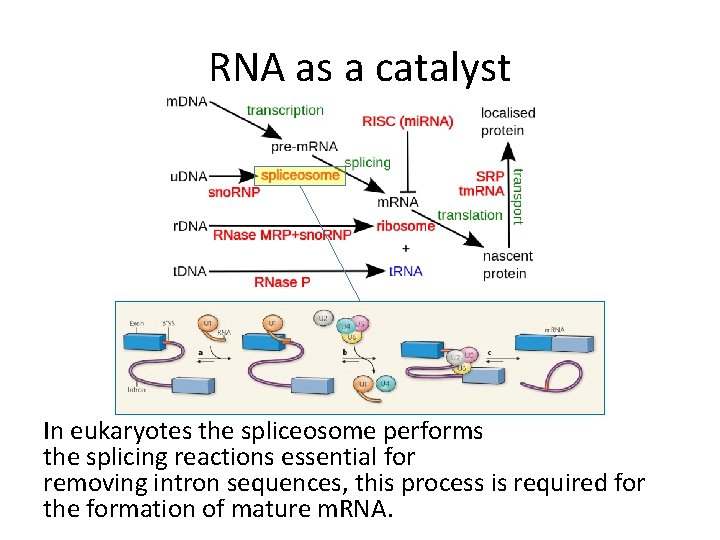
RNA role in organisms • The central dogma of biology: DNA RNA Protein Non Coding RNA • RNA as a catalyst: – Discovered in the early 1980’s.

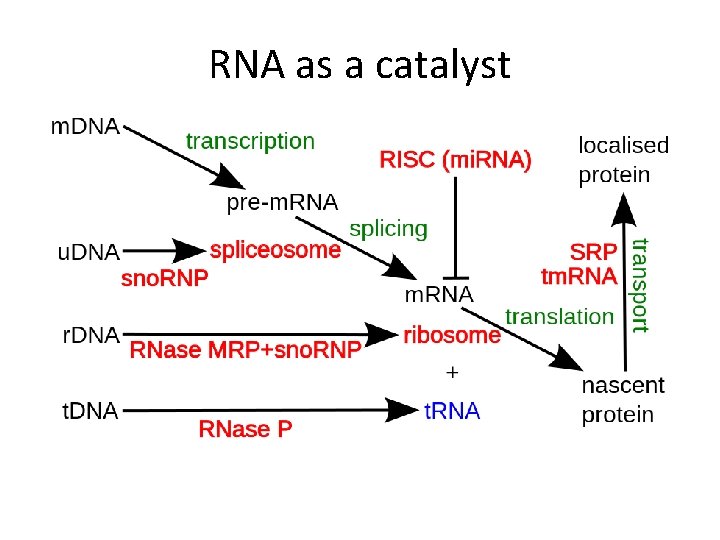
RNA as a catalyst

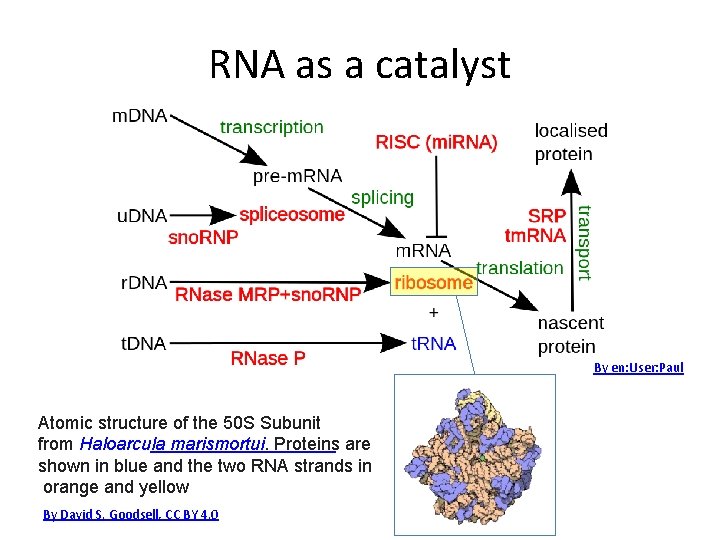
RNA as a catalyst By en: User: Paul Atomic structure of the 50 S Subunit from Haloarcula marismortui. Proteins are shown in blue and the two RNA strands in orange and yellow By David S. Goodsell, CC BY 4. 0

RNA as a catalyst In eukaryotes the spliceosome performs the splicing reactions essential for removing intron sequences, this process is required for the formation of mature m. RNA.

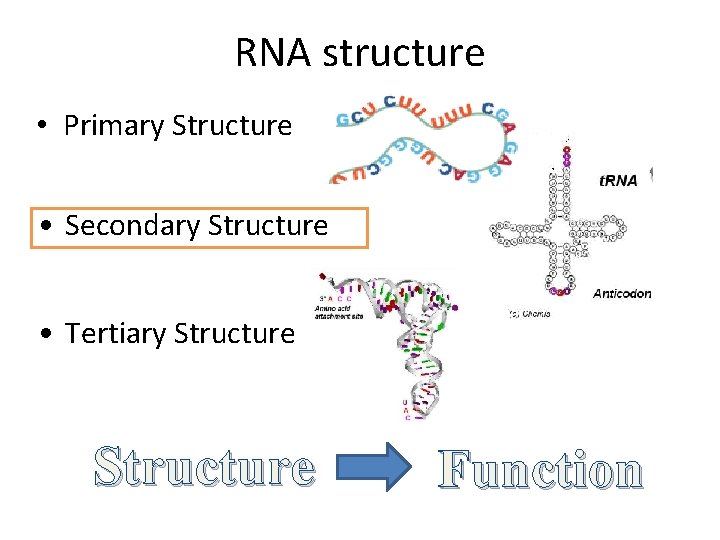
RNA structure • Primary Structure • Secondary Structure • Tertiary Structure Function

RNA secondary structure

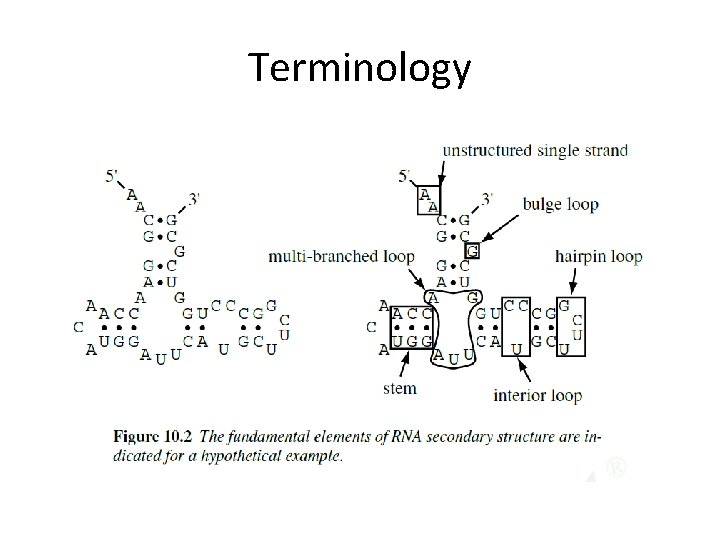
Terminology

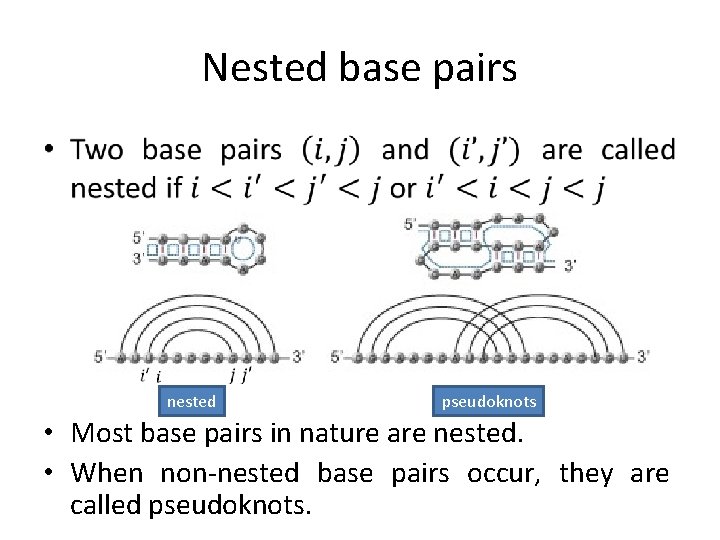
Nested base pairs • nested pseudoknots • Most base pairs in nature are nested. • When non-nested base pairs occur, they are called pseudoknots.

Sequence constrained by structure • It is relatively common to find examples of RNAs with a similar structure but without significant similarity. • It would be advantageous to be able to search for conserved structure in addition to sequence when searching similar RNAs.

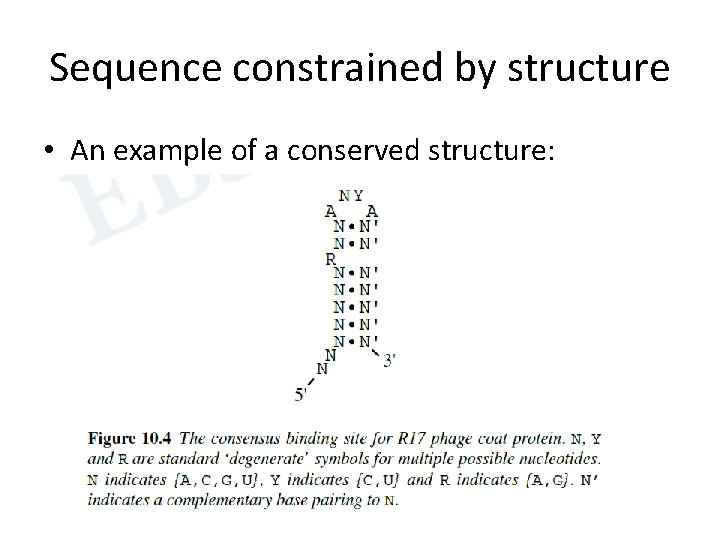
Sequence constrained by structure • An example of a conserved structure:

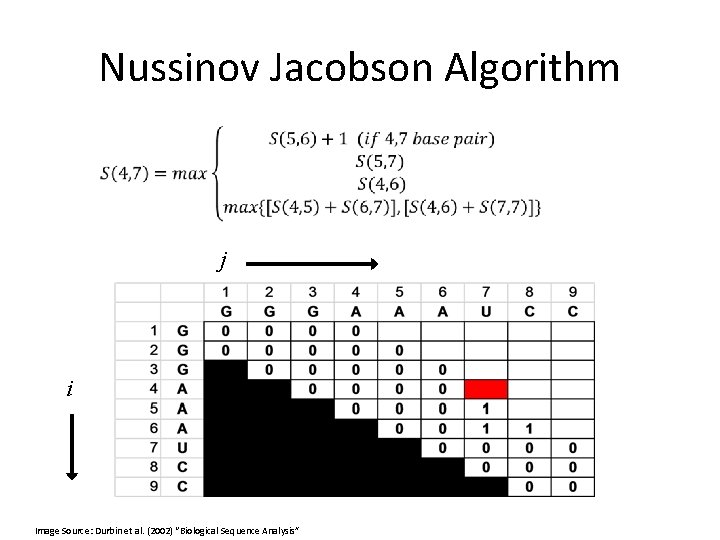
Nussinov Jacobson Algorithm • Ruth Nussinov proposed the first dynamic programming approach for RNA secondary structure prediction. • She is a Professor in the Department of Human Genetics, School of Medicine at Tel Aviv University.

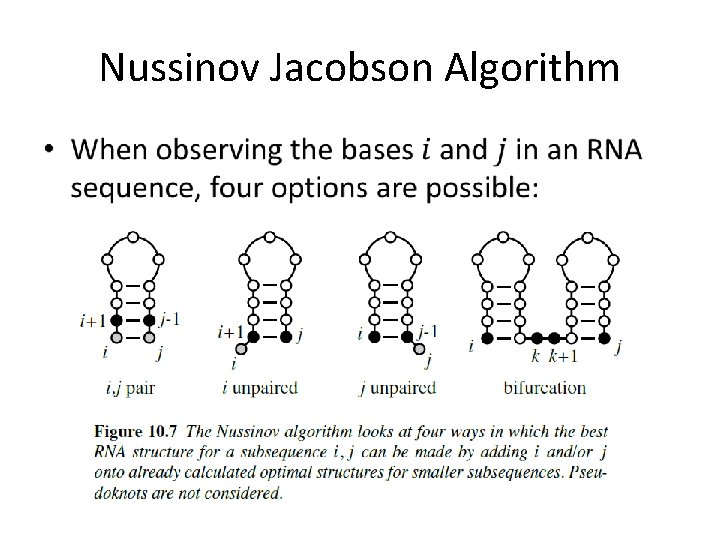
Nussinov Jacobson Algorithm •

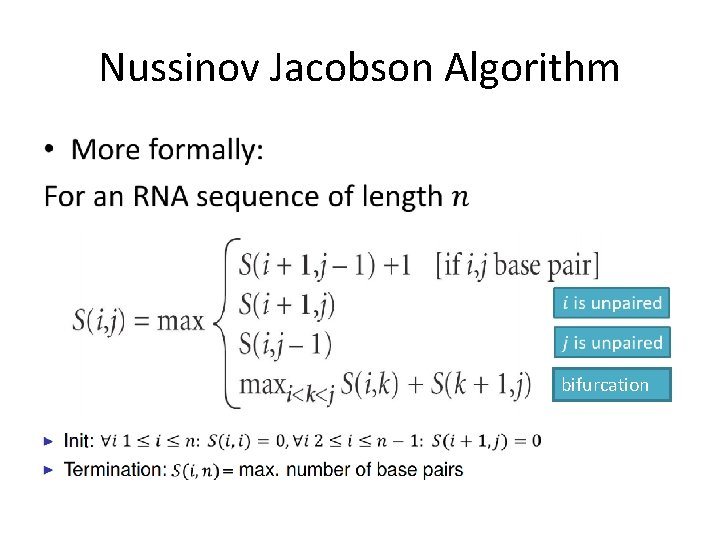
Nussinov Jacobson Algorithm The problem: • Find the secondary structure with the maximum number of paired bases Main idea: • Dynamic programming - the optimal solution to a certain sequence will be derived from the optimal solution of the sub-sequences

Nussinov Jacobson Algorithm • In order to solve our problem with DP, some assumptions must be made: • The assumptions: – No pseudoknots. – All base pairs are of equal energy. • Is this assumption necessary?

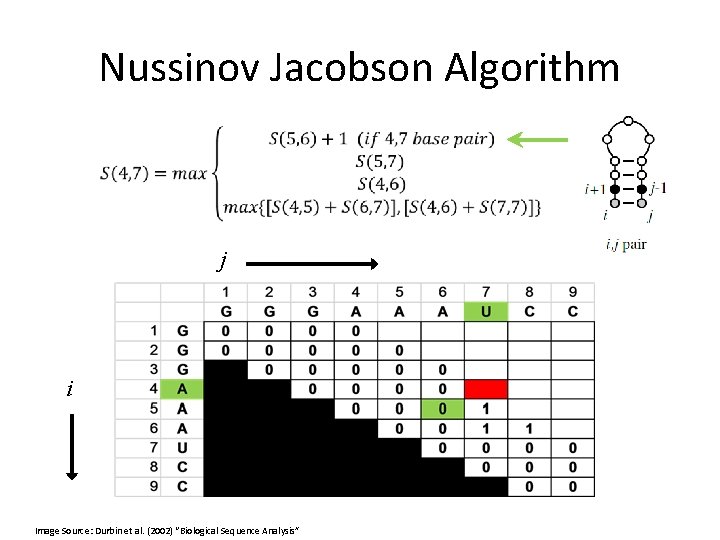
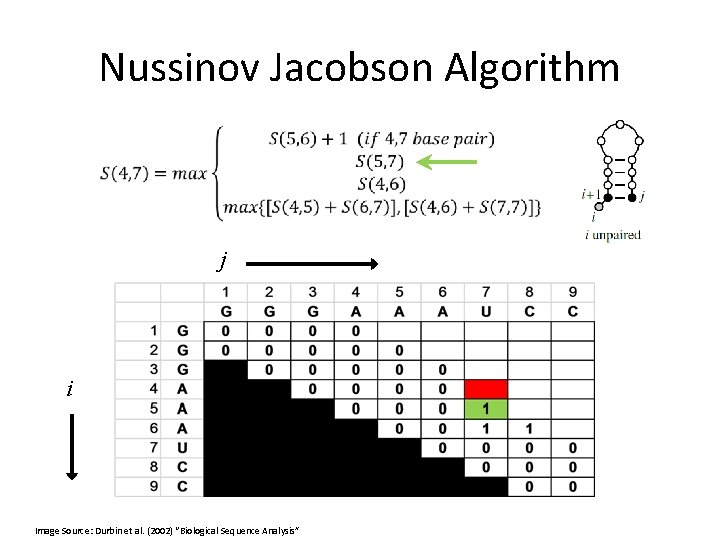
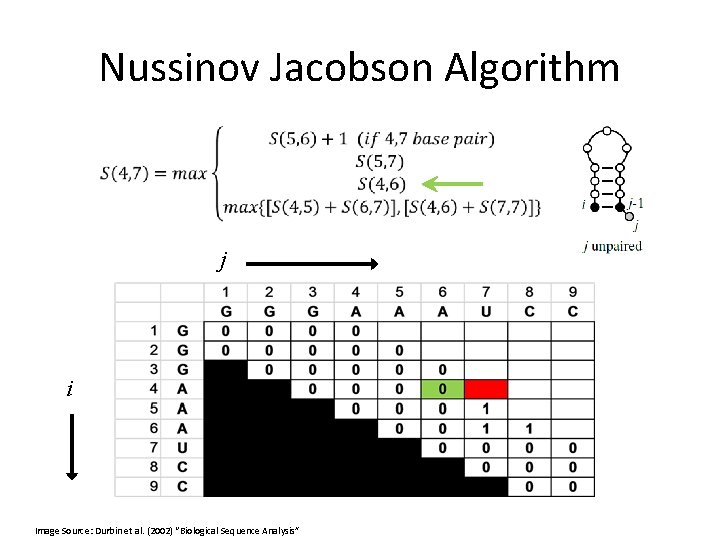
Nussinov Jacobson Algorithm •

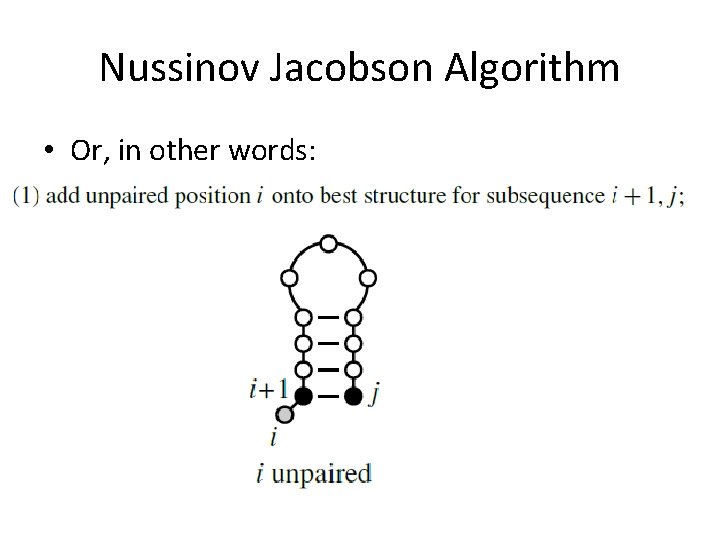
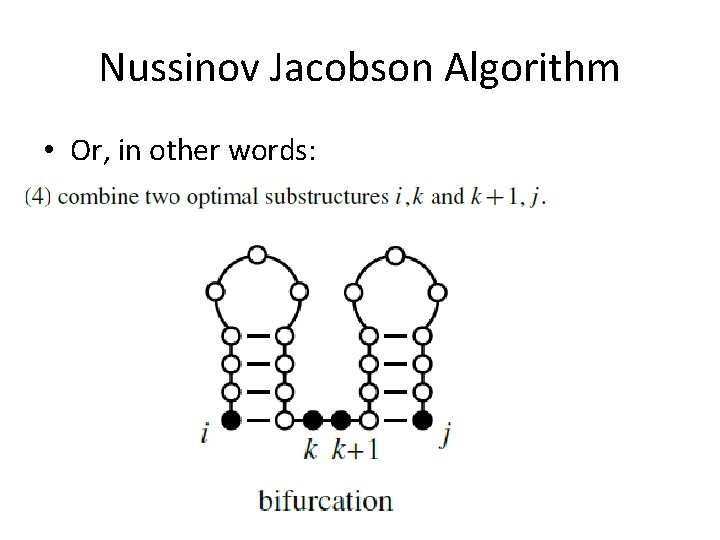
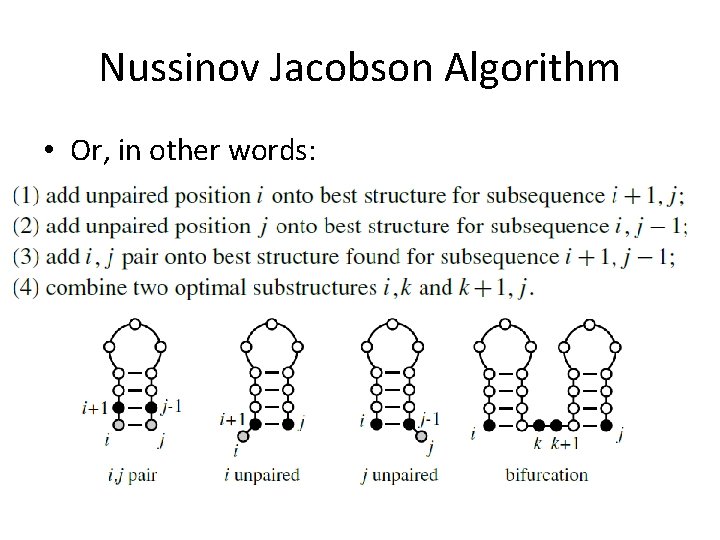
Nussinov Jacobson Algorithm • Or, in other words:

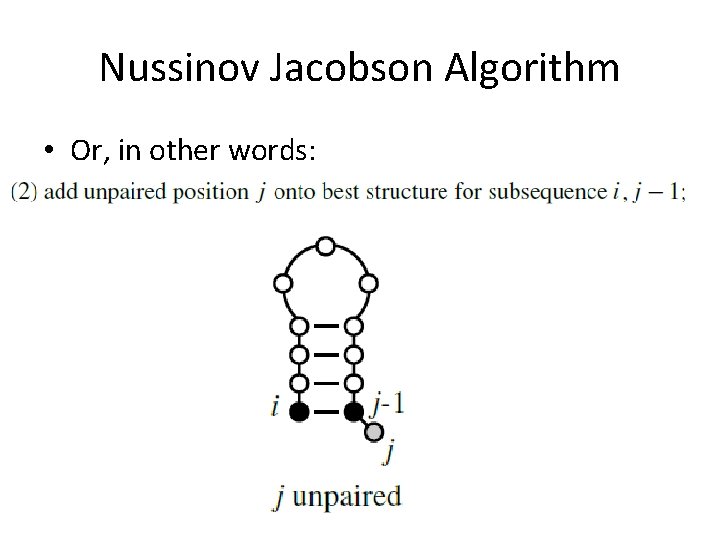
Nussinov Jacobson Algorithm • Or, in other words:

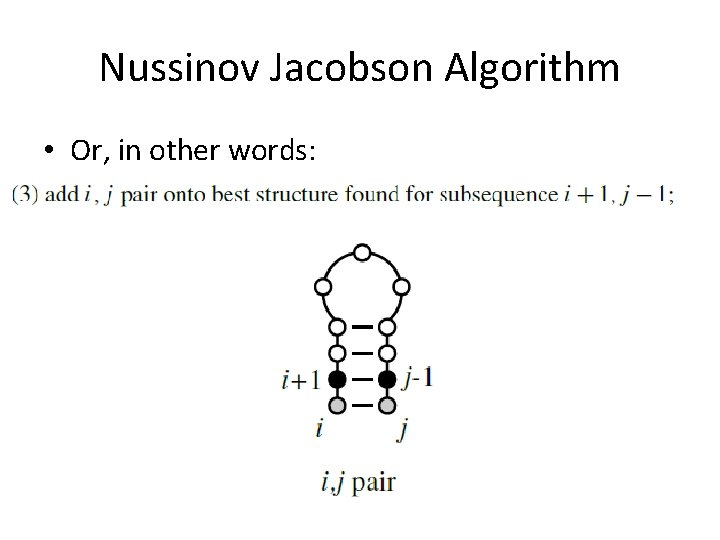
Nussinov Jacobson Algorithm • Or, in other words:

Nussinov Jacobson Algorithm • Or, in other words:

Nussinov Jacobson Algorithm • Or, in other words:

Nussinov Jacobson Algorithm • bifurcation

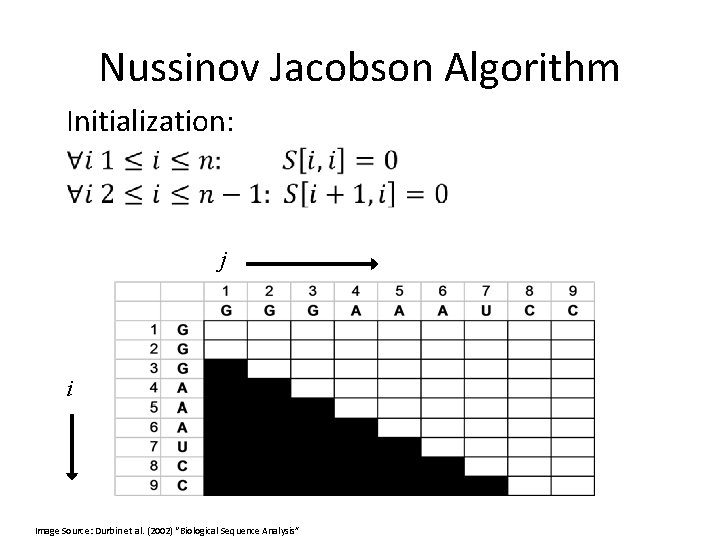
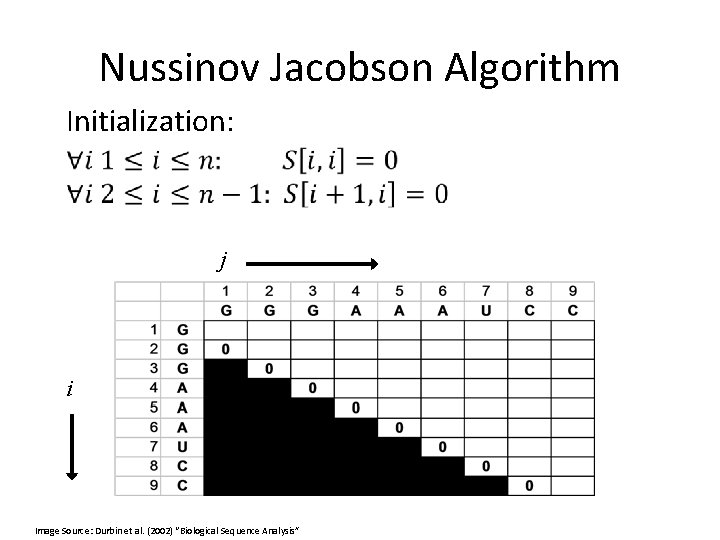
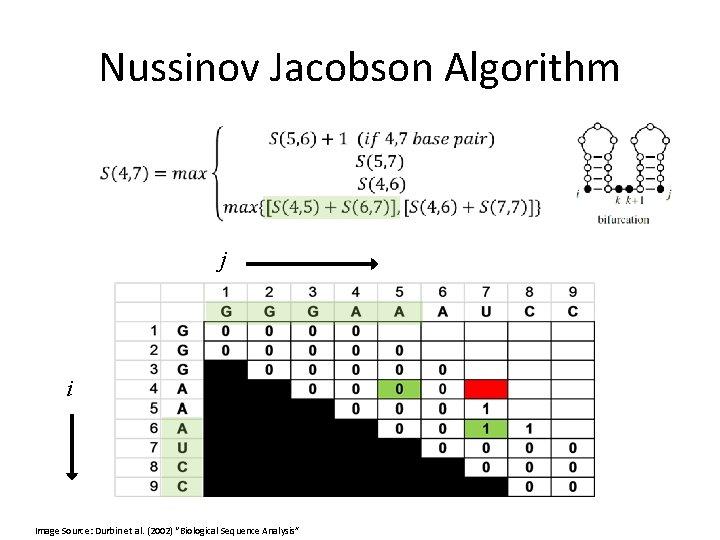
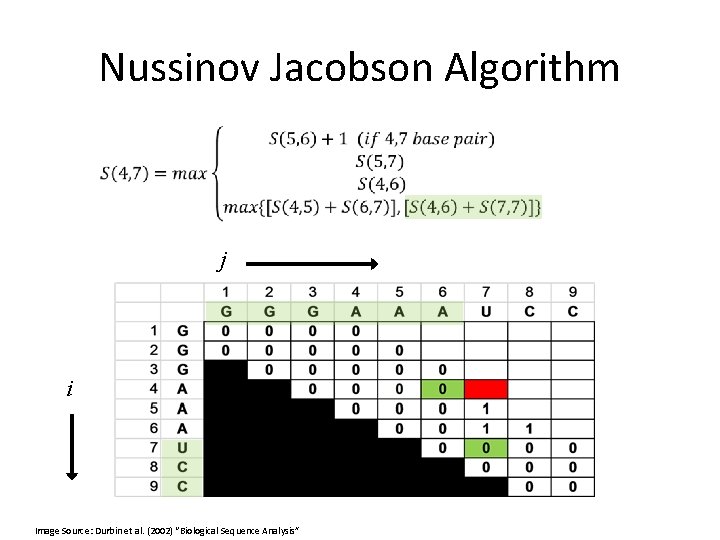
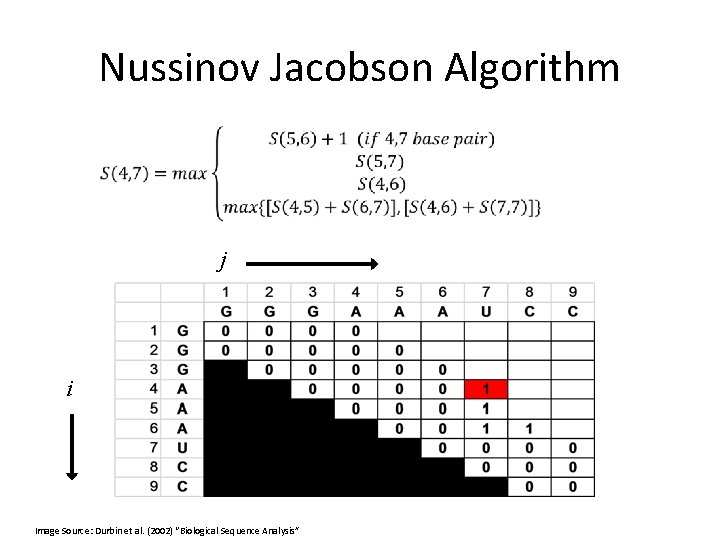
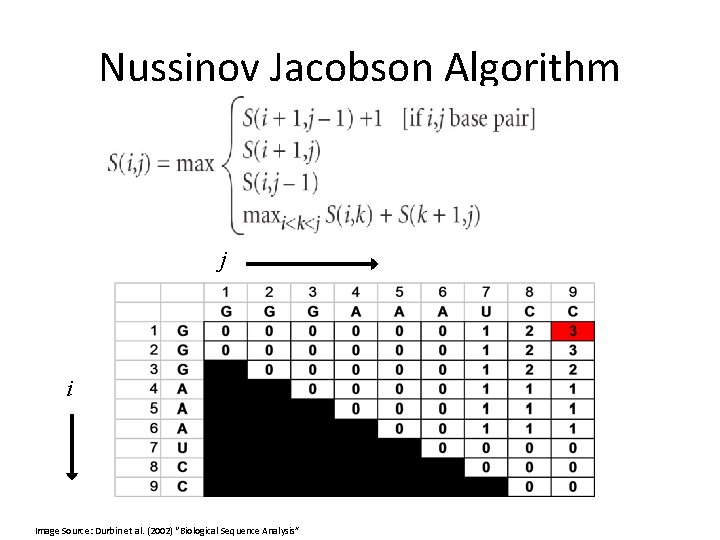
Nussinov Jacobson Algorithm Initialization: j i Image Source: Durbin et al. (2002) “Biological Sequence Analysis”

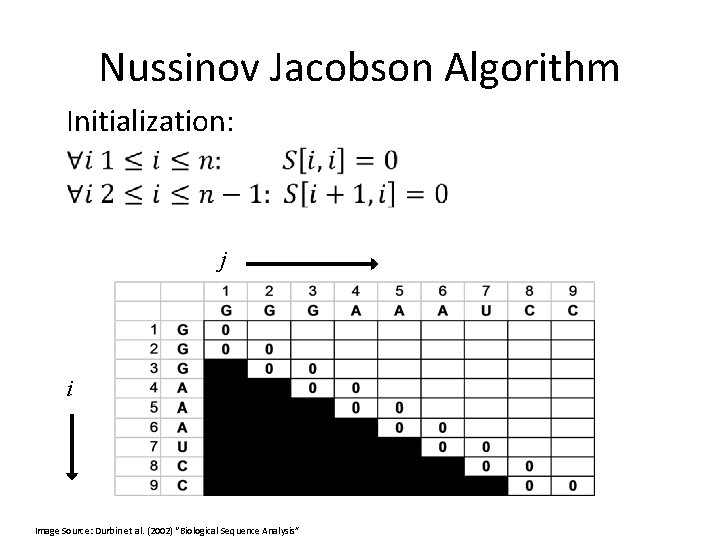
Nussinov Jacobson Algorithm Initialization: j i Image Source: Durbin et al. (2002) “Biological Sequence Analysis”

Nussinov Jacobson Algorithm Initialization: j i Image Source: Durbin et al. (2002) “Biological Sequence Analysis”

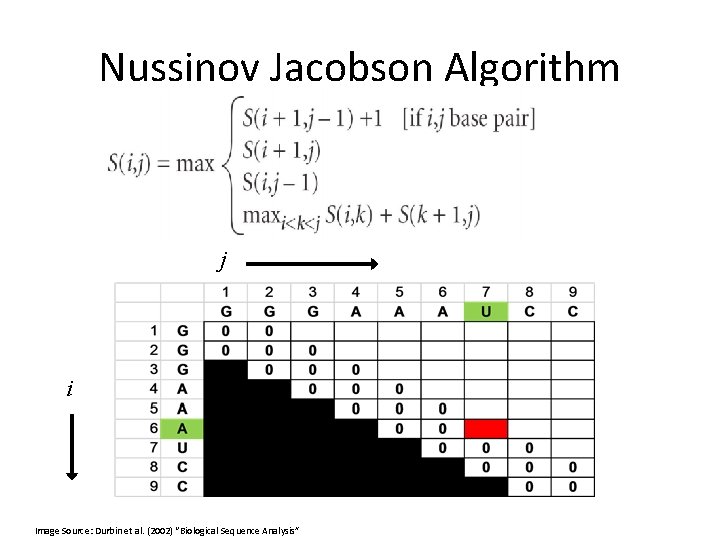
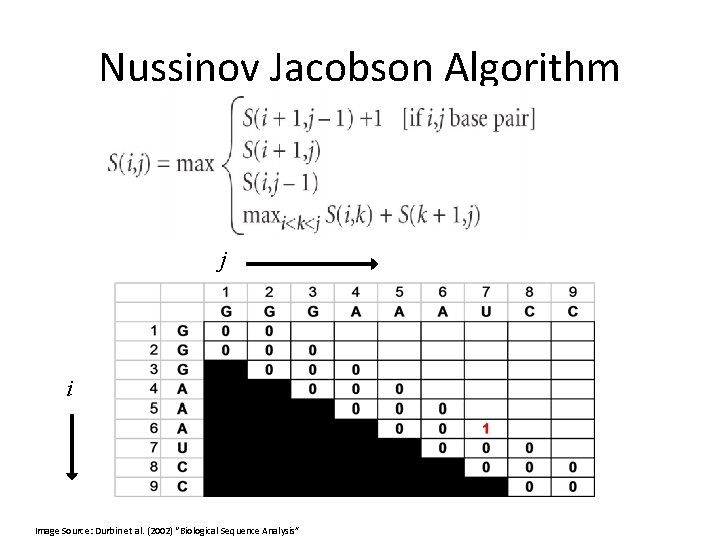
Nussinov Jacobson Algorithm j i Image Source: Durbin et al. (2002) “Biological Sequence Analysis”

Nussinov Jacobson Algorithm j i Image Source: Durbin et al. (2002) “Biological Sequence Analysis”

Nussinov Jacobson Algorithm j i Image Source: Durbin et al. (2002) “Biological Sequence Analysis”

Nussinov Jacobson Algorithm j i Image Source: Durbin et al. (2002) “Biological Sequence Analysis”

Nussinov Jacobson Algorithm j i Image Source: Durbin et al. (2002) “Biological Sequence Analysis”

Nussinov Jacobson Algorithm j i Image Source: Durbin et al. (2002) “Biological Sequence Analysis”

Nussinov Jacobson Algorithm j i Image Source: Durbin et al. (2002) “Biological Sequence Analysis”

Nussinov Jacobson Algorithm j i Image Source: Durbin et al. (2002) “Biological Sequence Analysis”

Nussinov Jacobson Algorithm j i Image Source: Durbin et al. (2002) “Biological Sequence Analysis”

Nussinov Jacobson Algorithm j i Image Source: Durbin et al. (2002) “Biological Sequence Analysis”

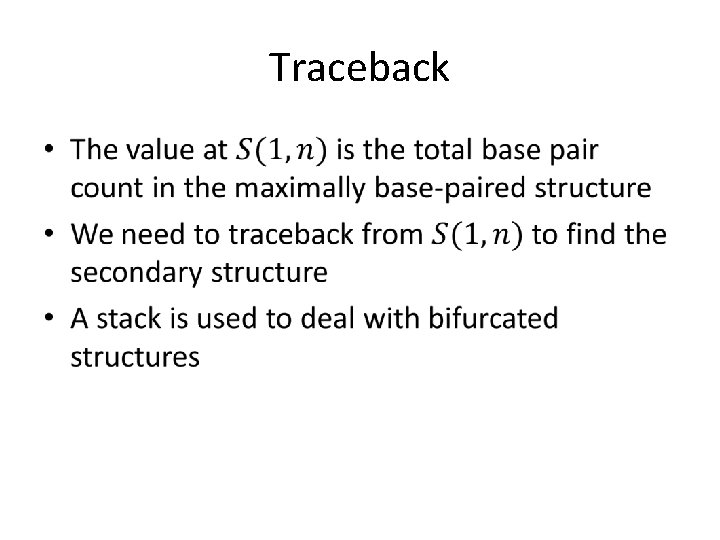
Traceback •

Traceback •

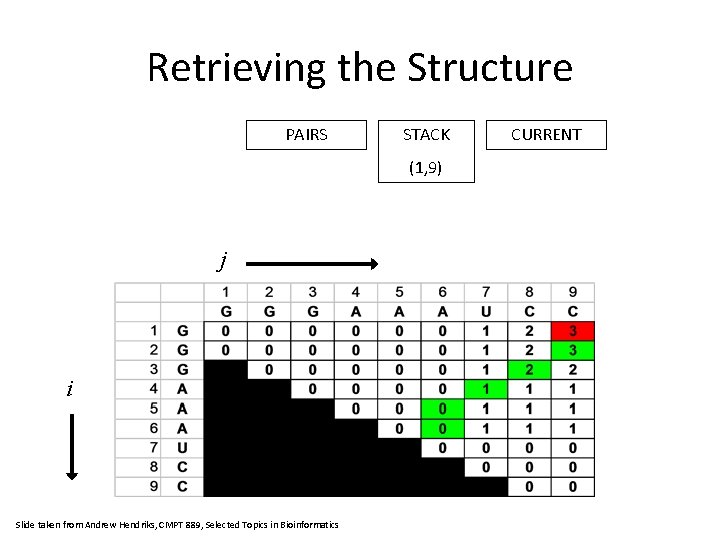
Retrieving the Structure PAIRS STACK (1, 9) j i Slide taken from Andrew Hendriks, CMPT 889, Selected Topics in Bioinformatics CURRENT

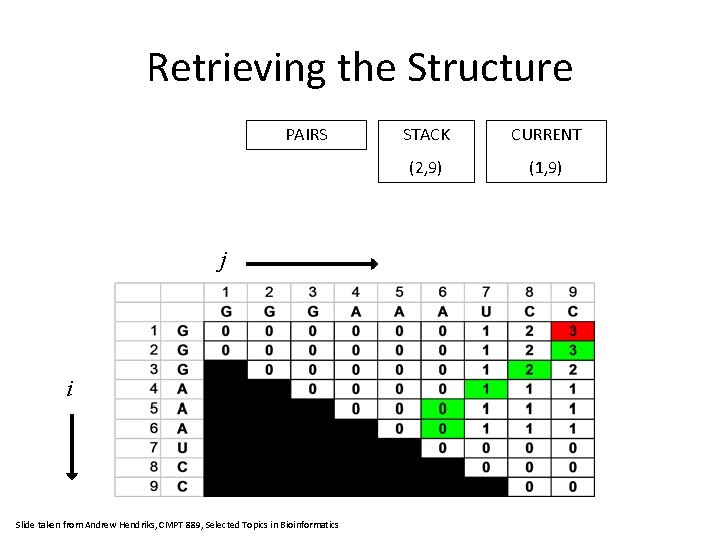
Retrieving the Structure PAIRS j i Slide taken from Andrew Hendriks, CMPT 889, Selected Topics in Bioinformatics STACK CURRENT (2, 9) (1, 9)

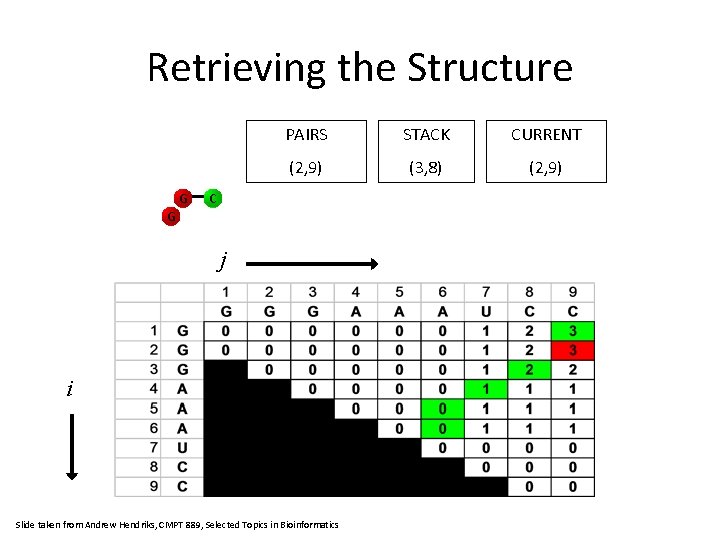
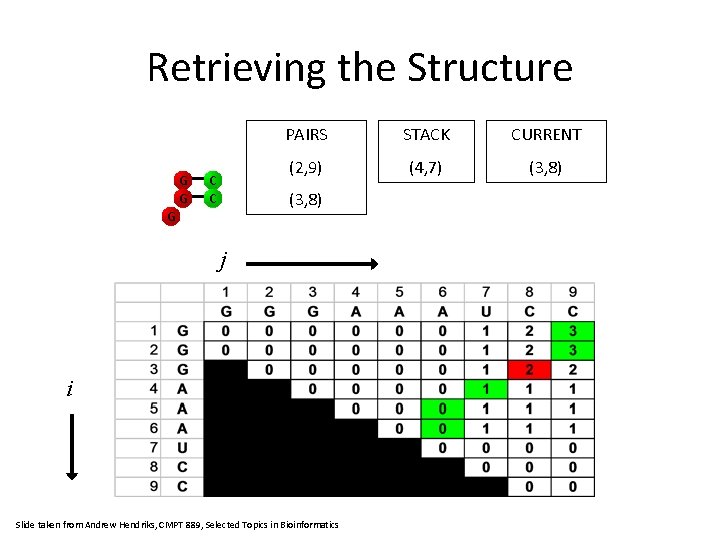
Retrieving the Structure G PAIRS STACK CURRENT (2, 9) (3, 8) (2, 9) C G j i Slide taken from Andrew Hendriks, CMPT 889, Selected Topics in Bioinformatics

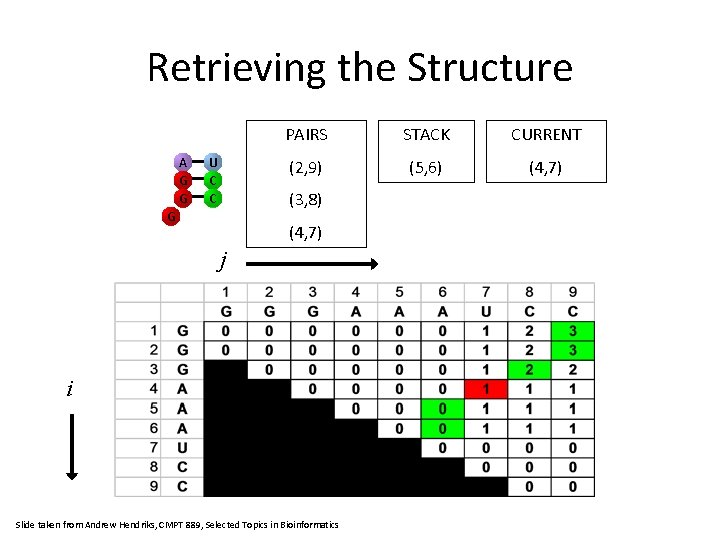
Retrieving the Structure G G C C PAIRS STACK CURRENT (2, 9) (4, 7) (3, 8) G j i Slide taken from Andrew Hendriks, CMPT 889, Selected Topics in Bioinformatics

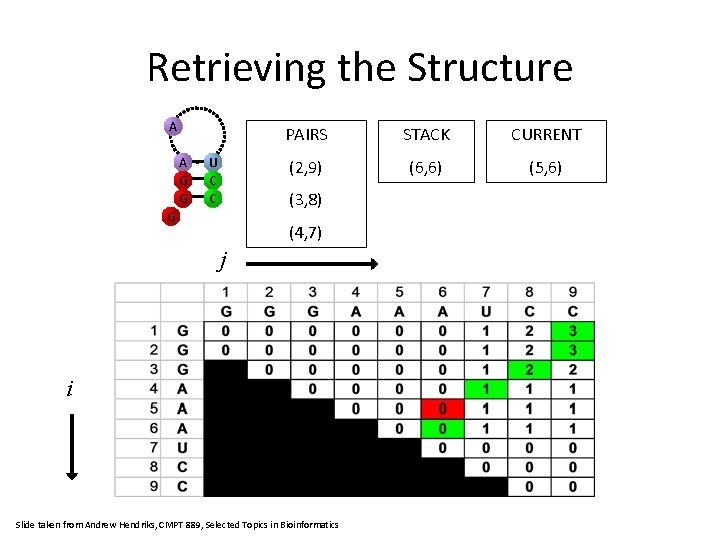
Retrieving the Structure A G G U C C PAIRS STACK CURRENT (2, 9) (5, 6) (4, 7) (3, 8) G (4, 7) j i Slide taken from Andrew Hendriks, CMPT 889, Selected Topics in Bioinformatics

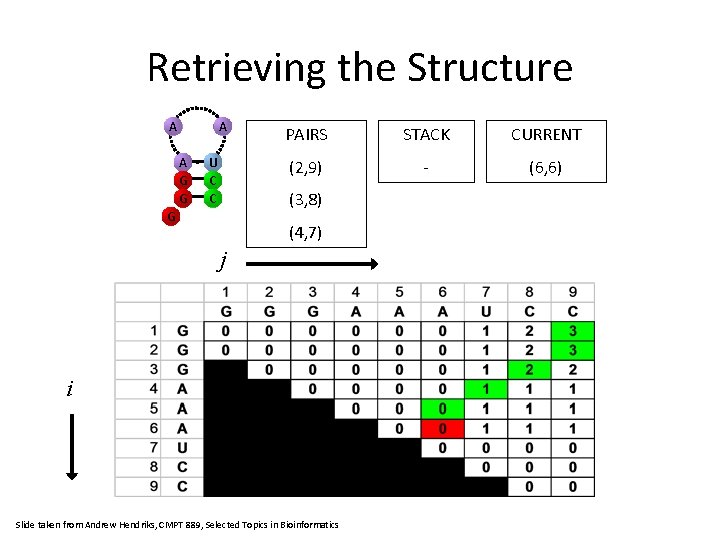
Retrieving the Structure A A G G U C C PAIRS STACK CURRENT (2, 9) (6, 6) (5, 6) (3, 8) G (4, 7) j i Slide taken from Andrew Hendriks, CMPT 889, Selected Topics in Bioinformatics

Retrieving the Structure A A A G G U C C PAIRS STACK CURRENT (2, 9) - (6, 6) (3, 8) G (4, 7) j i Slide taken from Andrew Hendriks, CMPT 889, Selected Topics in Bioinformatics

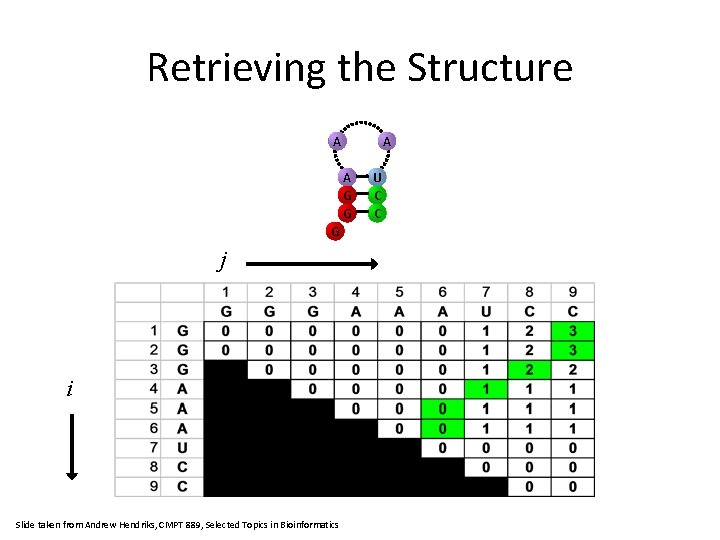
Retrieving the Structure A A A G G G j i Slide taken from Andrew Hendriks, CMPT 889, Selected Topics in Bioinformatics U C C

Nussinov Jacobson Algorithm •

Drawbacks • Doesn’t allow wobble base pairs like G-U • Doesn’t consider stacking: A-U A-U U-A A-U • Hair pin sizes not considered • More advanced algorithms consider energy minimization, to solve these issues.

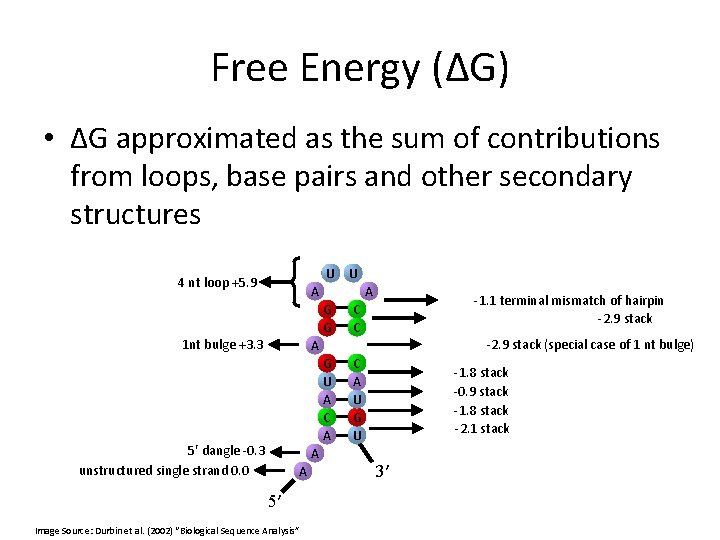
Free Energy (ΔG) • ΔG approximated as the sum of contributions from loops, base pairs and other secondary structures U U 4 nt loop +5. 9 A A G G 1 nt bulge +3. 3 -1. 1 terminal mismatch of hairpin -2. 9 stack C C -2. 9 stack (special case of 1 nt bulge) A G U A C A 5′ dangle -0. 3 unstructured single strand 0. 0 A A 5′ Image Source: Durbin et al. (2002) “Biological Sequence Analysis” C A U G U -1. 8 stack -0. 9 stack -1. 8 stack -2. 1 stack 3′