RNA secondary structure prediction Chase Beisel Tech talk

RNA secondary structure prediction Chase Beisel Tech talk 12 -21 -06
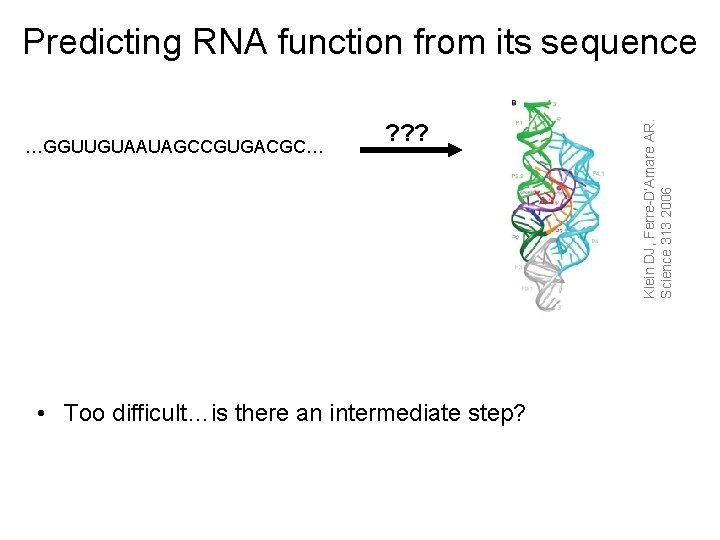
…GGUUGUAAUAGCCGUGACGC… ? ? ? • Too difficult…is there an intermediate step? Klein DJ, Ferre-D’Amare AR. Science 313 2006 Predicting RNA function from its sequence
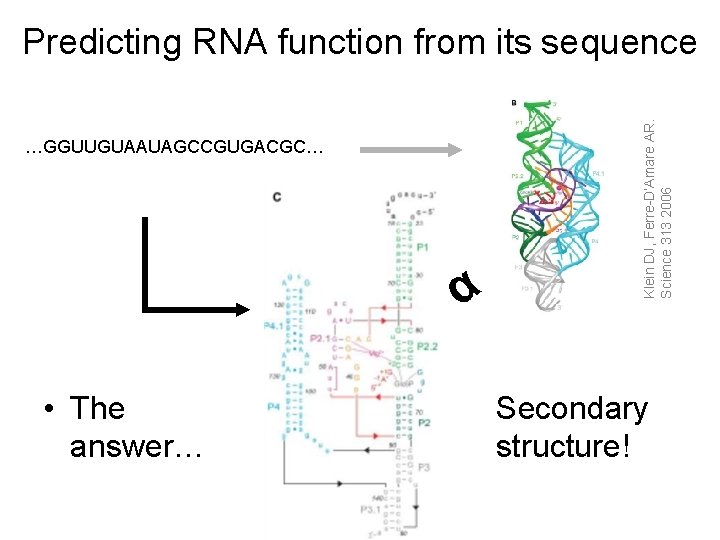
…GGUUGUAAUAGCCGUGACGC… α • The answer… Klein DJ, Ferre-D’Amare AR. Science 313 2006 Predicting RNA function from its sequence Secondary structure!
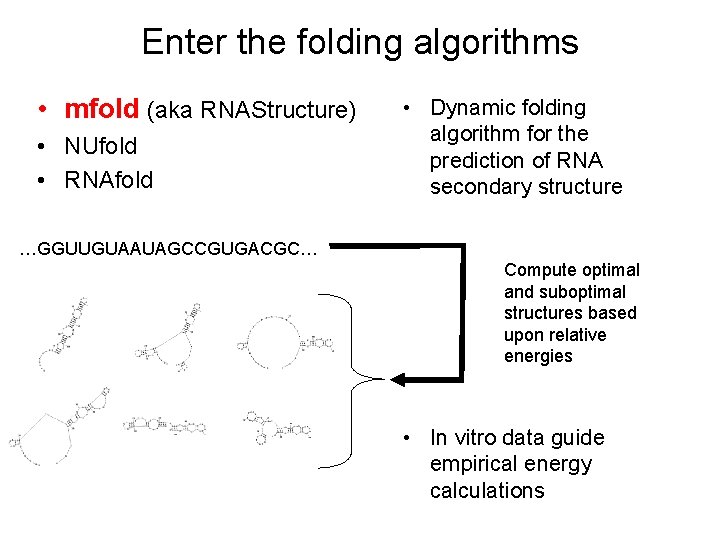
Enter the folding algorithms • mfold (aka RNAStructure) • NUfold • RNAfold • Dynamic folding algorithm for the prediction of RNA secondary structure …GGUUGUAAUAGCCGUGACGC… Compute optimal and suboptimal structures based upon relative energies • In vitro data guide empirical energy calculations

Enter the folding algorithms • mfold (aka RNAStructure) • NUfold • RNAfold • Dynamic folding algorithm for the prediction of RNA secondary structure …GGUUGUAAUAGCCGUGACGC… Compute optimal and suboptimal structures based upon relative energies • In vitro data guide empirical energy calculations

Explaining the available parameters
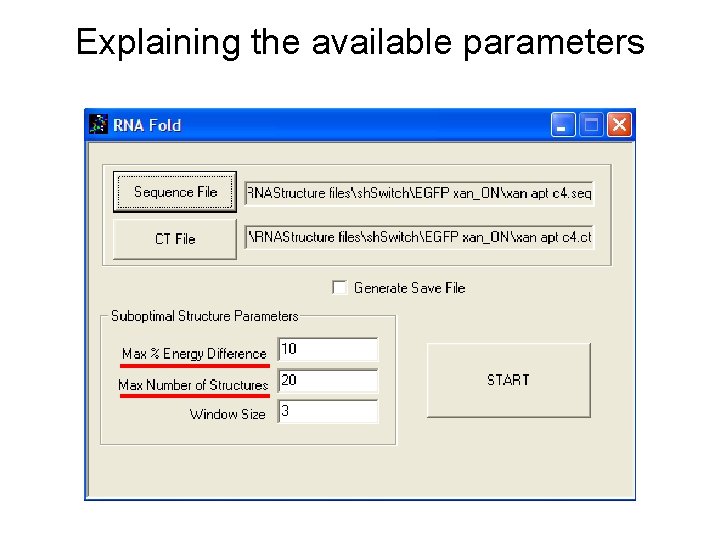
Explaining the available parameters
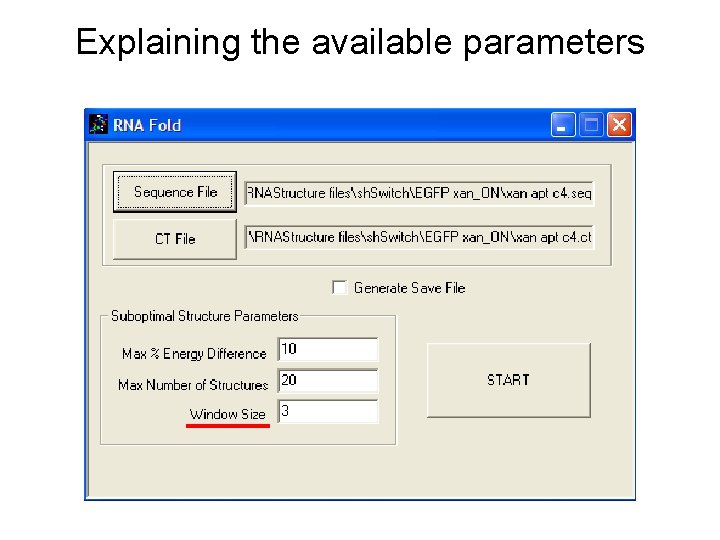
Explaining the available parameters
Caveats of mfold, or the problem with in vivo application • Cannot predict formation of pseudoknots nor anything beyond canonical and wobble base pair interactions% • In vitro data collected under standardized conditions: – 1 M Na. Cl, 37 ºC, no divalent ions@ • Assumes equilibrium conditions# • Only 67%* of known base pairs are correctly predicted by the optimal structure – 84. 4%* predicted by the best suboptimal structure % Eddy SR. Nature Biotechnol 22(11) 2004 # @ Draper DE. RNA 10 2004 Grilley D et al. PNAS 103(38) 2006 * Mathews DH, et al. PNAS 101(19) 2004 Pan T & Sosnick T. ARBBS 35 2006
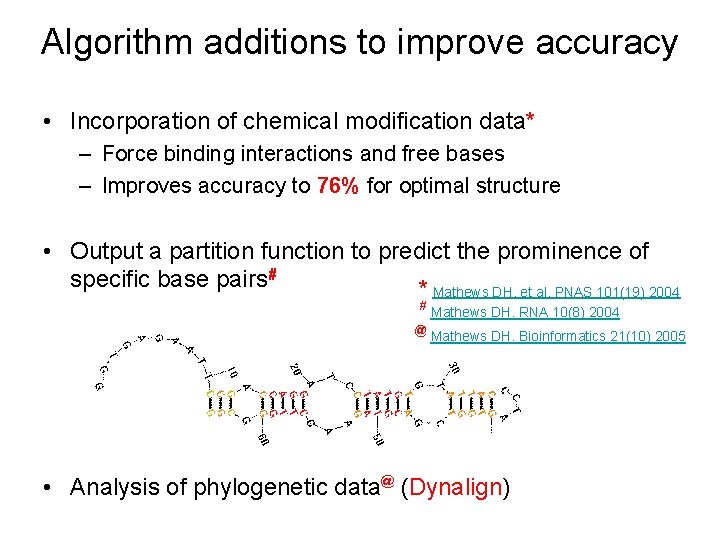
Algorithm additions to improve accuracy • Incorporation of chemical modification data* – Force binding interactions and free bases – Improves accuracy to 76% for optimal structure • Output a partition function to predict the prominence of specific base pairs# * Mathews DH, et al. PNAS 101(19) 2004 # @ Mathews DH. RNA 10(8) 2004 Mathews DH. Bioinformatics 21(10) 2005 • Analysis of phylogenetic data@ (Dynalign)
The take home message… …GGUUGUAAGC… ∆G = • Don’t treat RNAStructure like a black box!
- Slides: 11