RNA maturation transport localization RNA export to the

- Slides: 20

RNA maturation, transport & localization RNA export to the cytoplasm: model systems RNA degradation RNA localization in the cytoplasm

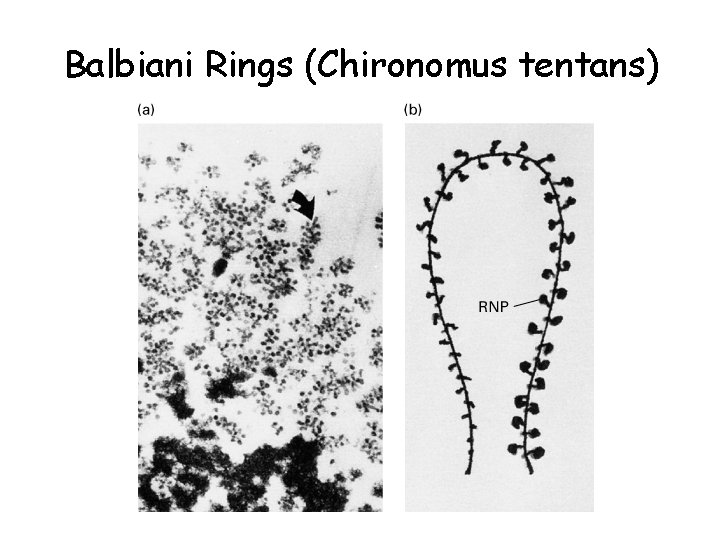
Balbiani Rings (Chironomus tentans)

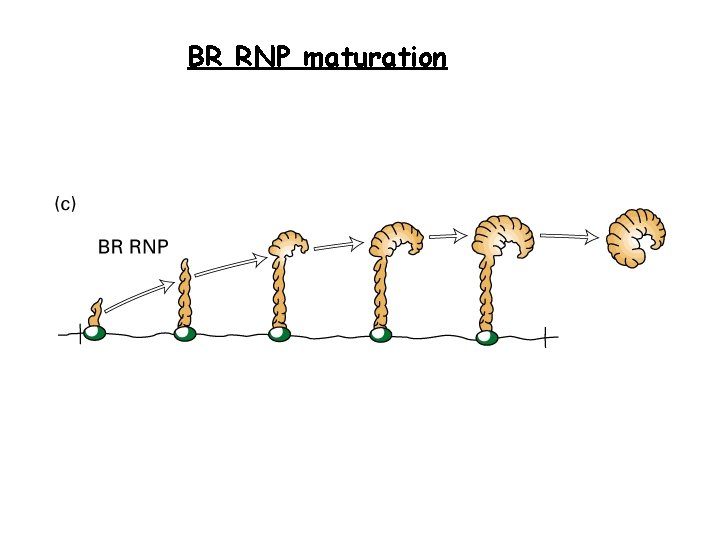
BR RNP maturation

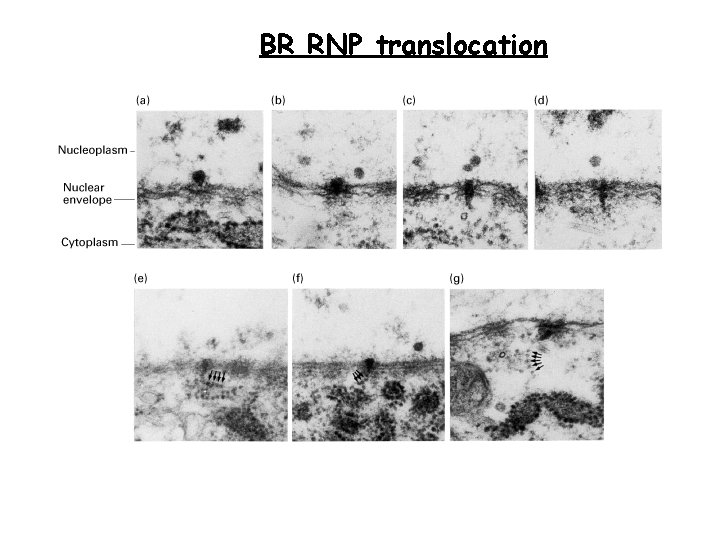
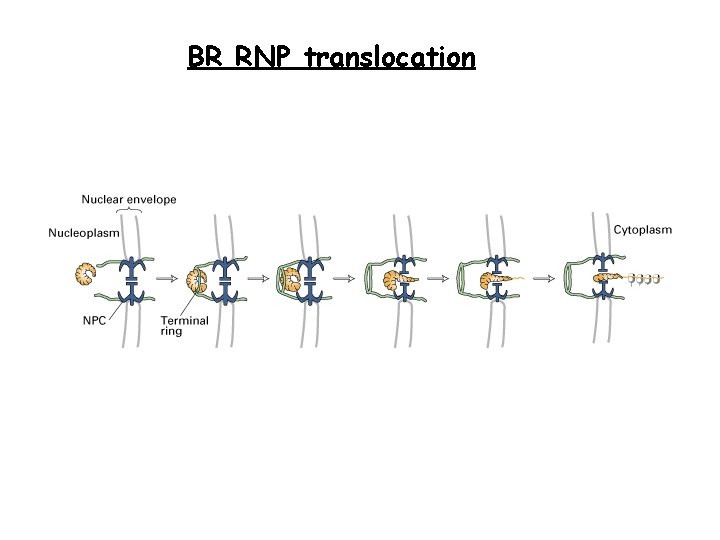
BR RNP translocation

BR RNP translocation


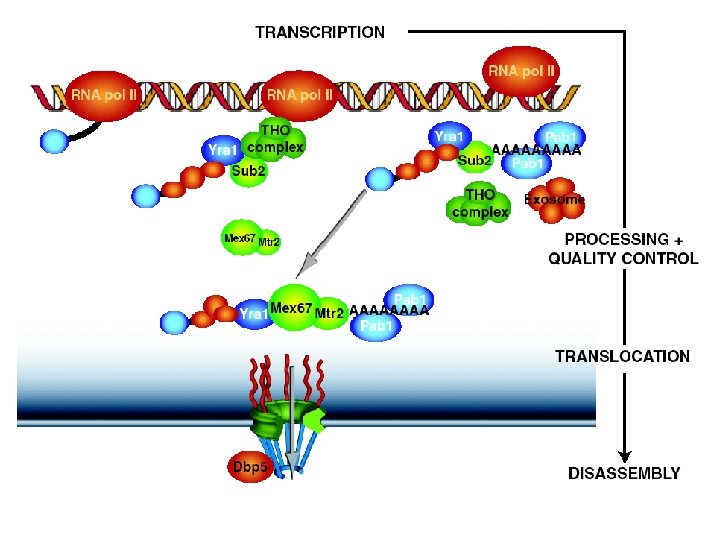
SUMMARY I. Multiple steps of m. RNA biosynthesis are tightly coupled II. Mex 67/TAP is one major m. RNA export factor Binding to m. RNA may already occur at the site of transcription III. Many questions remain - How are m. RNA substrates released in the cytoplasm? - Are there multiple m. RNA export pathways? - How is processing and transport coupled? etc.

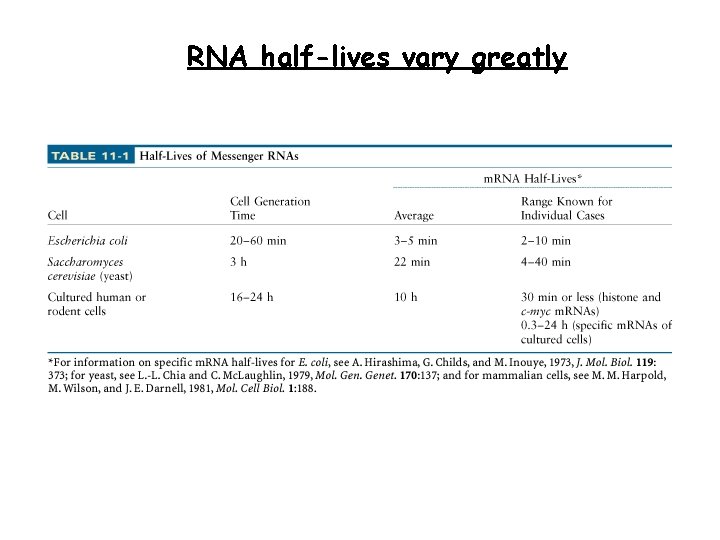
RNA half-lives vary greatly

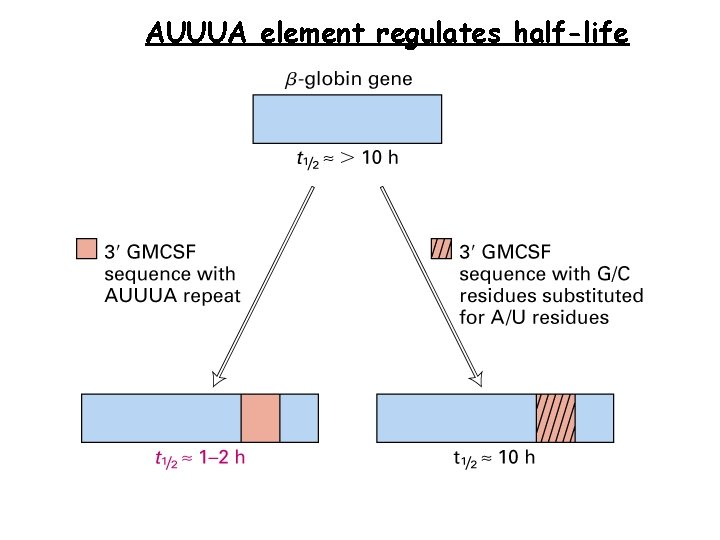
AUUUA element regulates half-life

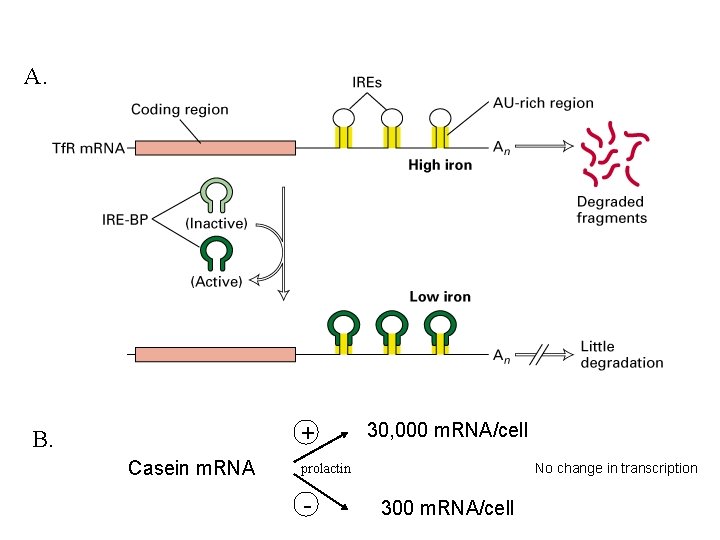
A. + B. Casein m. RNA 30, 000 m. RNA/cell No change in transcription prolactin - 300 m. RNA/cell

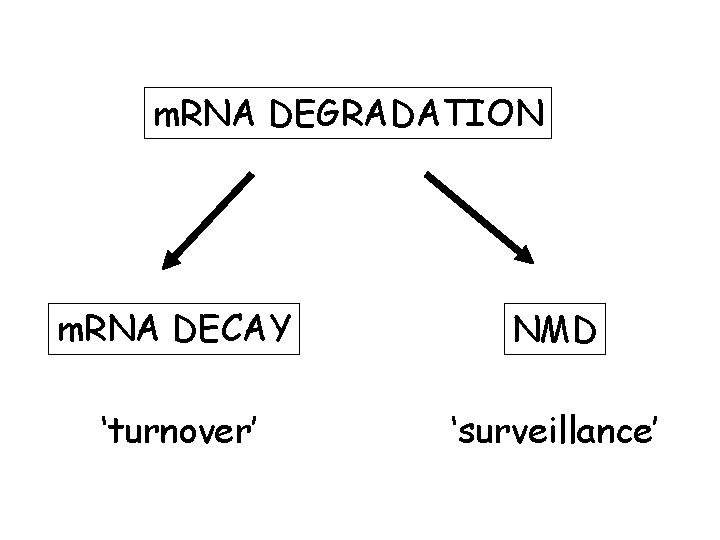
m. RNA DEGRADATION m. RNA DECAY NMD ‘turnover’ ‘surveillance’

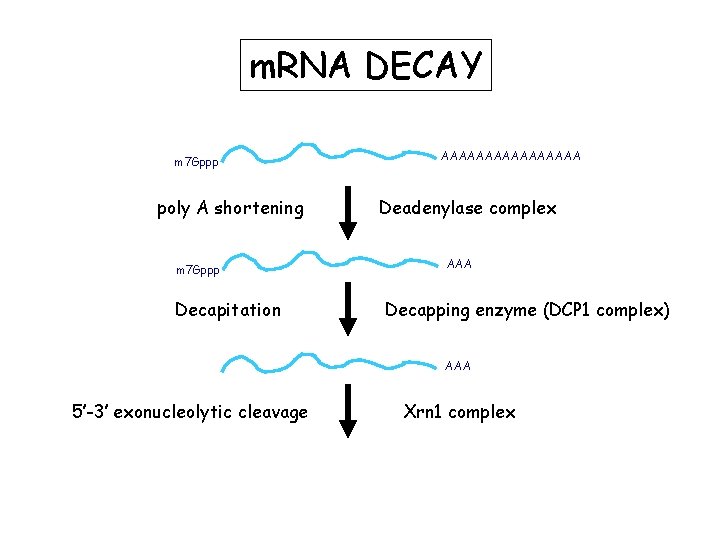
m. RNA DECAY m 7 Gppp poly A shortening m 7 Gppp Decapitation AAAAAAAA Deadenylase complex AAA Decapping enzyme (DCP 1 complex) AAA 5’-3’ exonucleolytic cleavage Xrn 1 complex

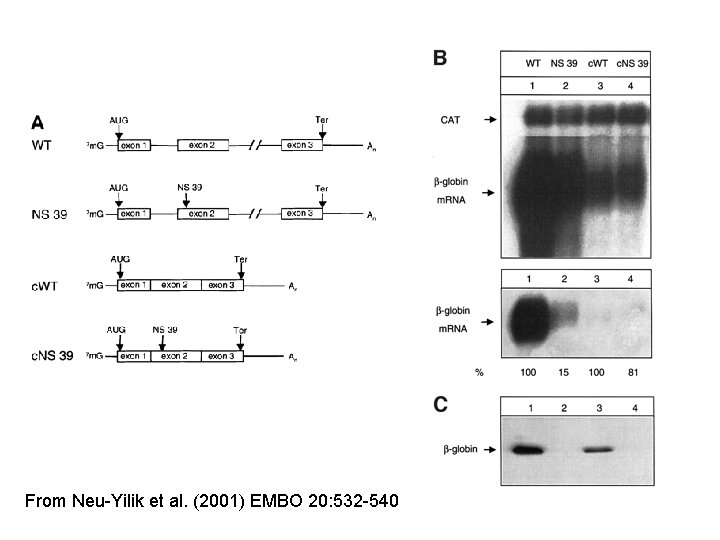
From Neu-Yilik et al. (2001) EMBO 20: 532 -540

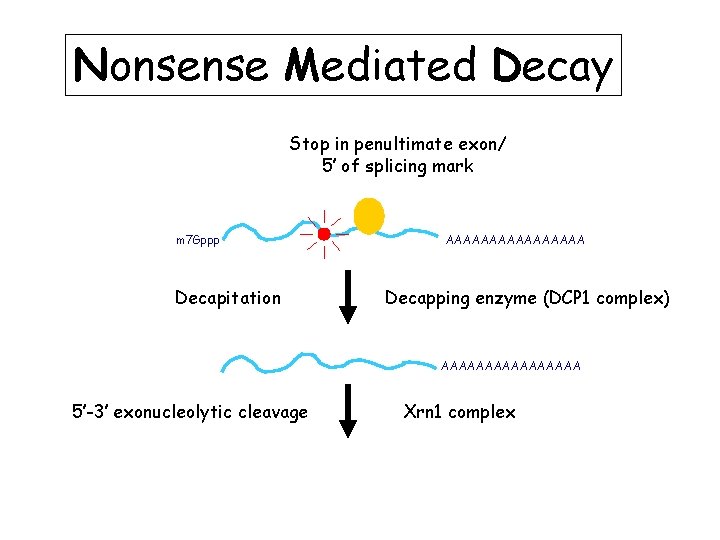
Nonsense Mediated Decay Stop in penultimate exon/ 5’ of splicing mark m 7 Gppp Decapitation AAAAAAAA Decapping enzyme (DCP 1 complex) AAAAAAAA 5’-3’ exonucleolytic cleavage Xrn 1 complex

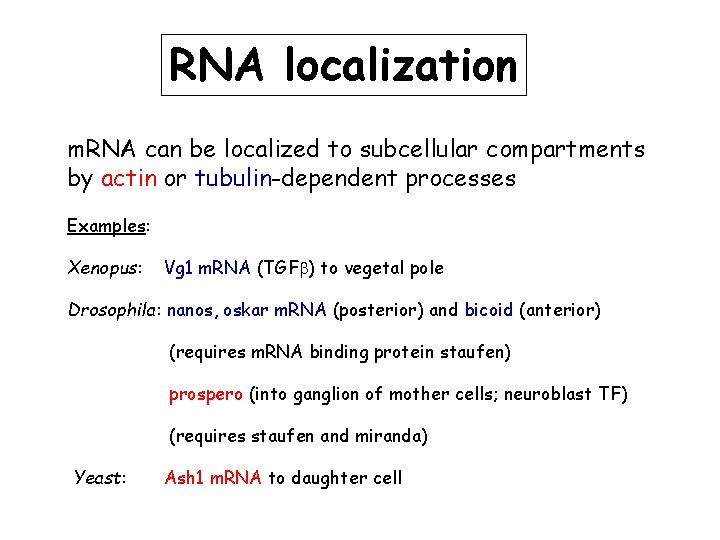
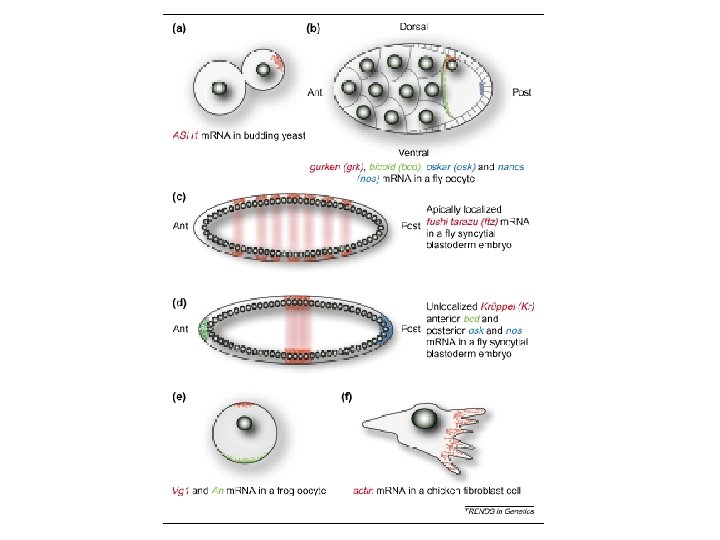
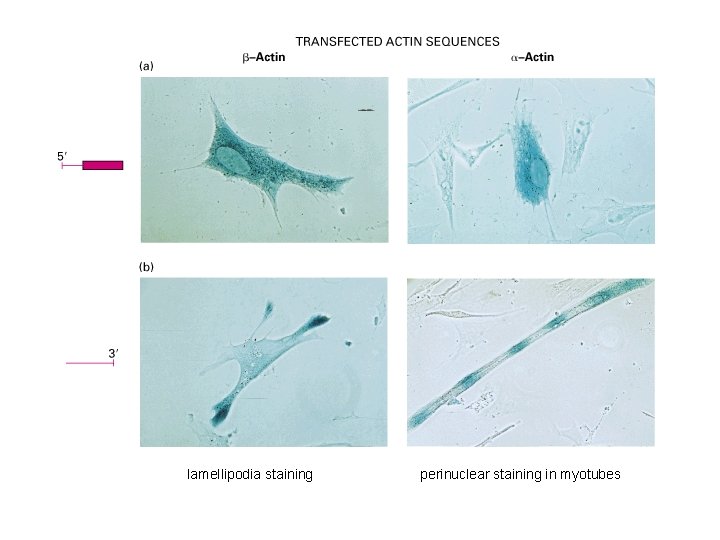
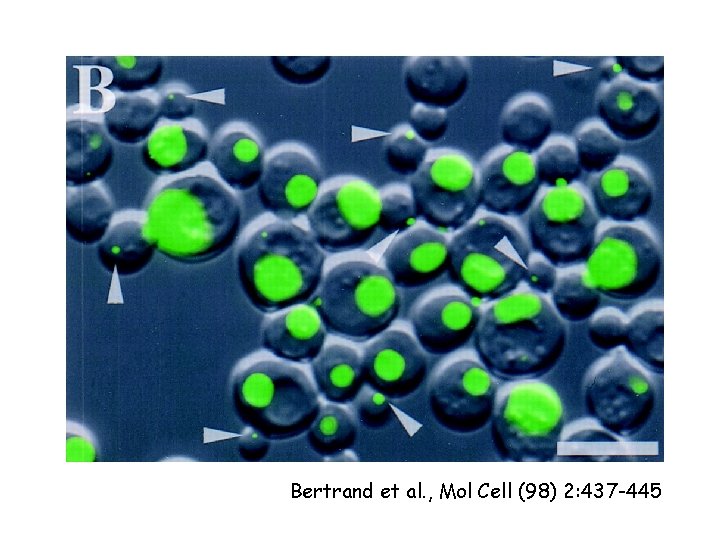
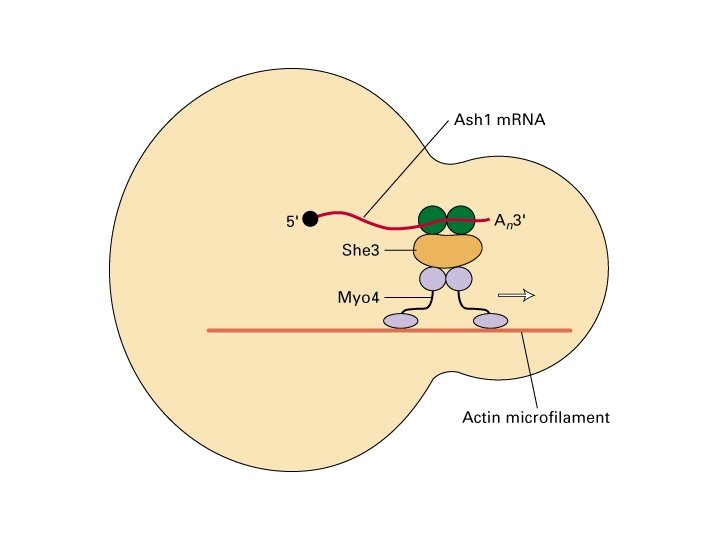
RNA localization m. RNA can be localized to subcellular compartments by actin or tubulin-dependent processes Examples: Xenopus: Vg 1 m. RNA (TGFb) to vegetal pole Drosophila: nanos, oskar m. RNA (posterior) and bicoid (anterior) (requires m. RNA binding protein staufen) prospero (into ganglion of mother cells; neuroblast TF) (requires staufen and miranda) Yeast: Ash 1 m. RNA to daughter cell


lamellipodia staining perinuclear staining in myotubes

Bertrand et al. , Mol Cell (98) 2: 437 -445


SUMMARY 2 I. m. RNA decay - regulated and non-regulated turn-over - ordered pathway (deadenylation, decapping, exonucleolytic degradation) - NMD: recognition of premature stop codons II. Cytoplasmic m. RNA localization - ZIP code in 3’ UTR - both actin and tubulin-mediated - yeast mating type switch as a model: Ash 1 m. RNA localization (via 3’ UTR, She 2/3, Myo 4 and actin cables)