RNA Lab Isolation quantification and q PCR analysis
RNA Lab (Isolation, quantification and q. PCR analysis) MCB 7300
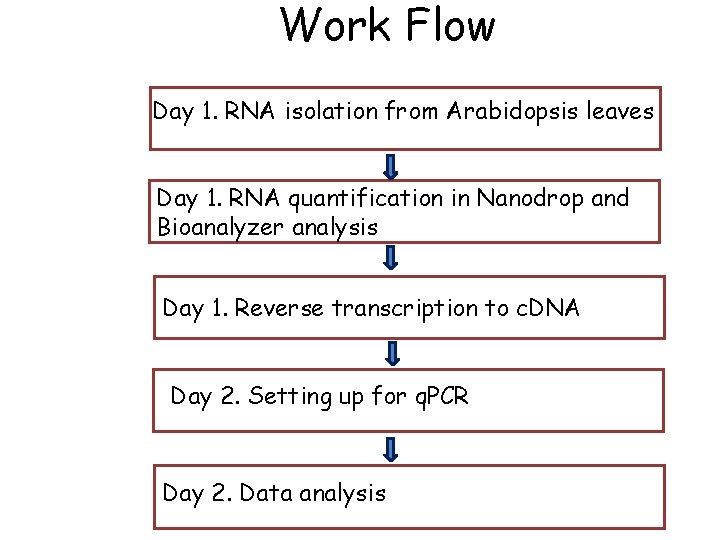
Work Flow Day 1. RNA isolation from Arabidopsis leaves Day 1. RNA quantification in Nanodrop and Bioanalyzer analysis Day 1. Reverse transcription to c. DNA Day 2. Setting up for q. PCR Day 2. Data analysis

Method of evaluation • You will have an in class quiz worth 50 points on Wednesday (Feb. 18 th) and a lab report worth 50 points which must be turned in by next Wednesday (i. e. , Feb. 25). • Please submit a hard copy of your lab report.
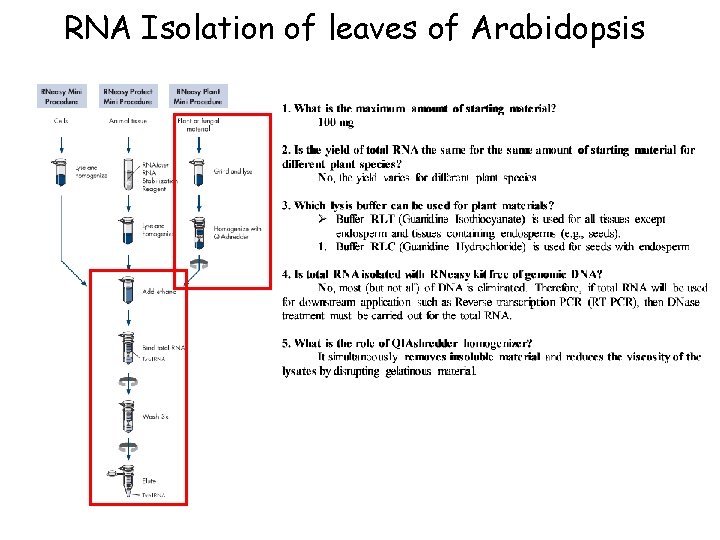
RNA Isolation of leaves of Arabidopsis

Quality check for RNA samples • UV/VIS Ratios – Absorption 260/230 ratio ≥ 1. 0 and 260/280 ratio ≥ 1. 8 – Low 260/280 ratios are often attributed to phenol and/or protein contaminations. – Low 260/230 ratios are usually attributed to salt (e. g. guanidine isothiocyanate) and/or phenol contaminations. – “High-salt”, seen as 260/230 ratio less than 1. 0, • Bio-analyzer – RIN (RNA integrity) ranges from 1 to 10, with 1 being the most degraded profile and 10 being the most intact. • Gel Electrophoresis – RNA sample integrity can also be evaluated using one of several standard denaturing gel electrophoresis methods.

Quantification of RNA samples by Nanodrop
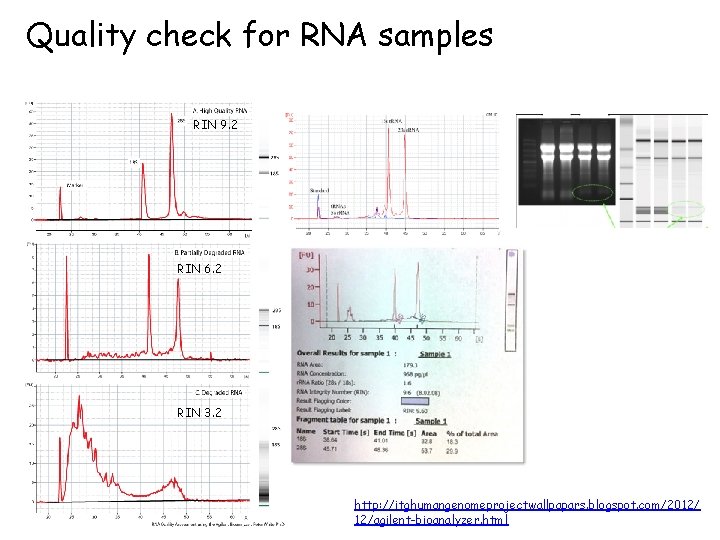
Quality check for RNA samples RIN 9. 2 RIN 6. 2 RIN 3. 2 http: //itghumangenomeprojectwallpapars. blogspot. com/2012/ 12/agilent-bioanalyzer. html

Real-Time q. PCR • Real‐time q. PCR is the most sensitive and reliable method for detection and quantification of nucleic acids (DNA, c. DNA, & RNA) levels. • It is based on detection and quantification of fluorescence. • Emitted from a reporter molecule at real time. • This detection occurs during the accumulation of the PCR product with each cycle of amplification, thus allows monitoring the PCR reaction during early & exponential phase where the first significant increase in the amount of PCR product correlates to the initial amount of target template.
Applications of q. PCR § § § § § Gene Expression Profiling Analysis Microarray Validation mi. RNA Expression Profiling Analysis Gene Regulation ‐‐‐ Genetic & Epigenetic SNP Genotyping & allelic discrimination Somatic Mutation Analysis Copy Number Detection/Variation Analysis Pathogen Detection Viral Quantification

Considerations • Isolation of m. RNA from total RNA (oligo d. T) or random hexamer primers • Choice of primers • Amplicon size and GC content
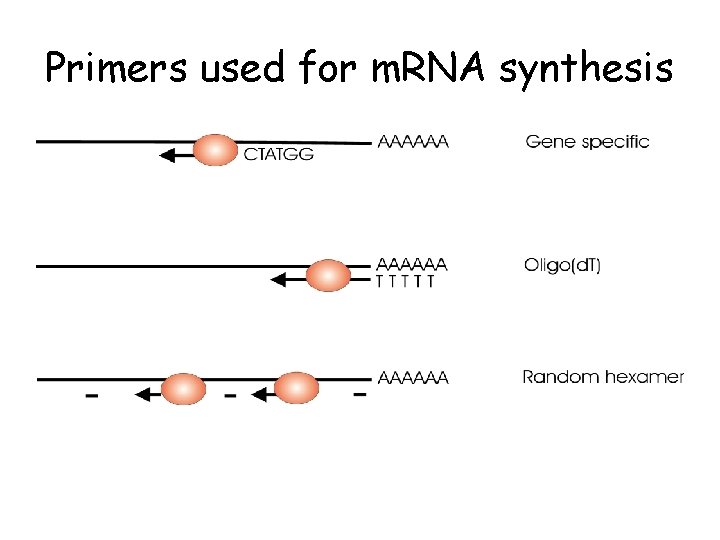
Primers used for m. RNA synthesis
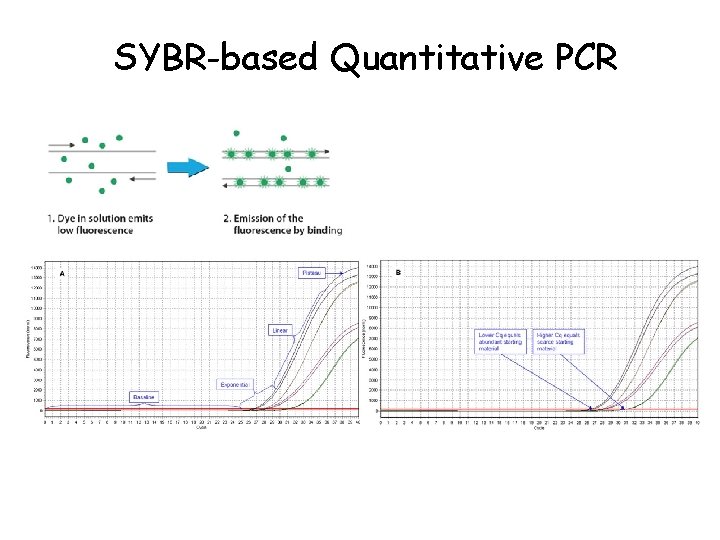
SYBR-based Quantitative PCR
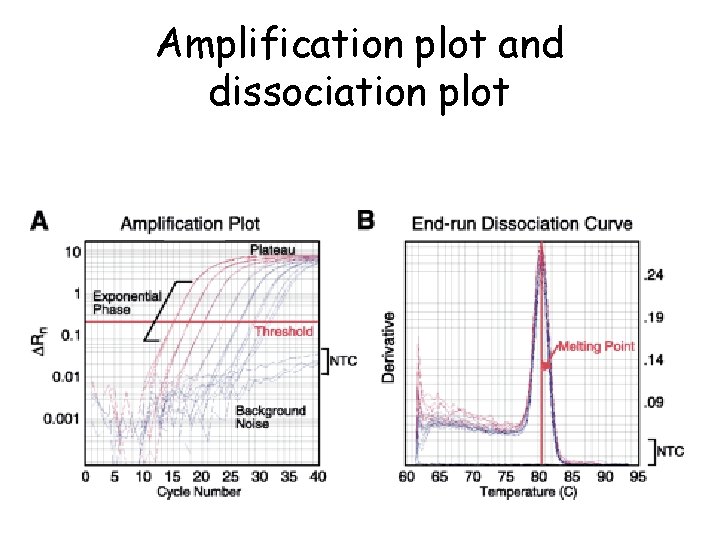
Amplification plot and dissociation plot
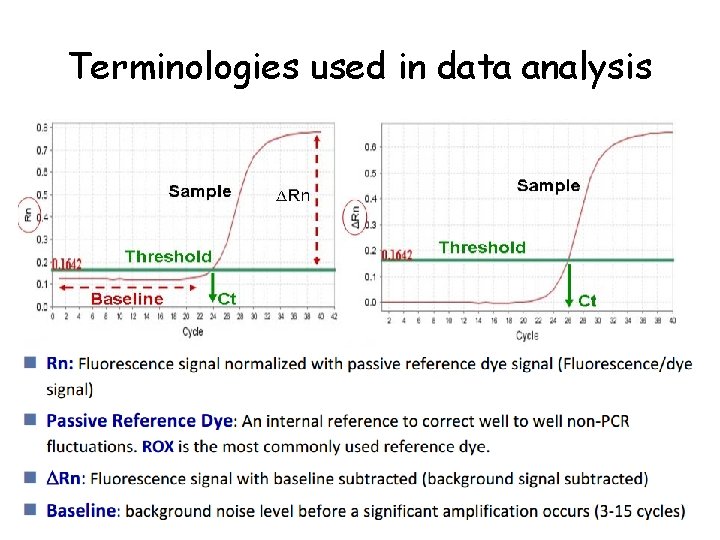
Terminologies used in data analysis
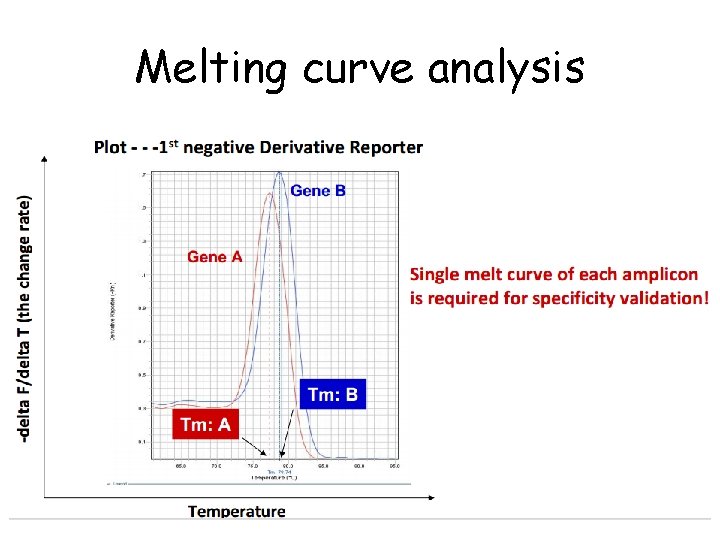
Melting curve analysis
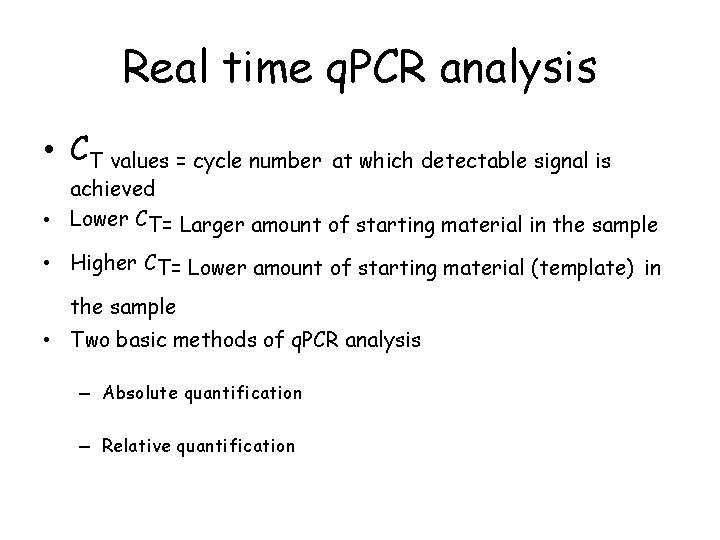
Real time q. PCR analysis • CT values = cycle number at which detectable signal is achieved • Lower CT= Larger amount of starting material in the sample • Higher CT= Lower amount of starting material (template) in the sample • Two basic methods of q. PCR analysis – Absolute quantification – Relative quantification
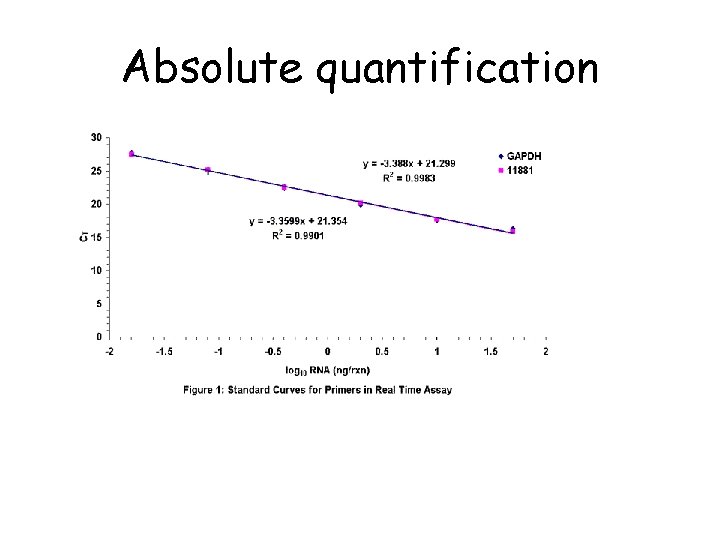
Absolute quantification
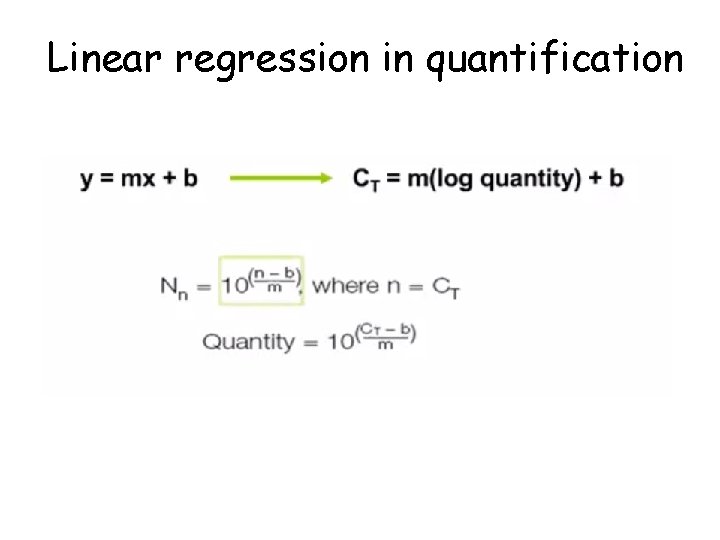
Linear regression in quantification

Applications • Viral load determination • Gene copy number determination
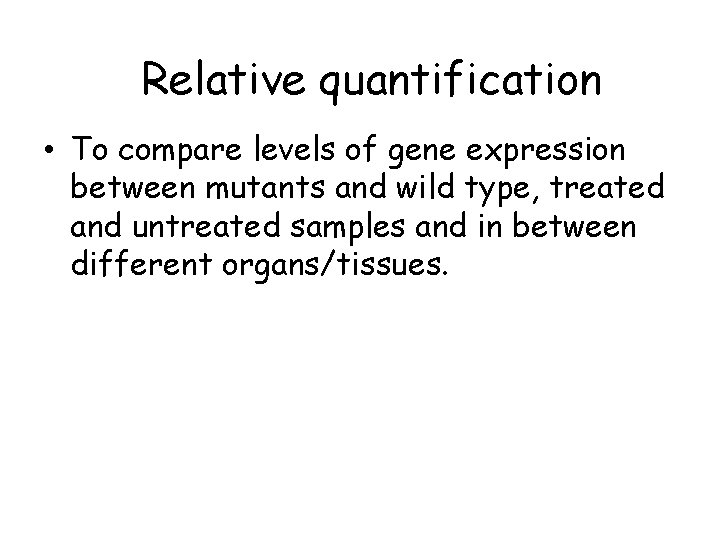
Relative quantification • To compare levels of gene expression between mutants and wild type, treated and untreated samples and in between different organs/tissues.
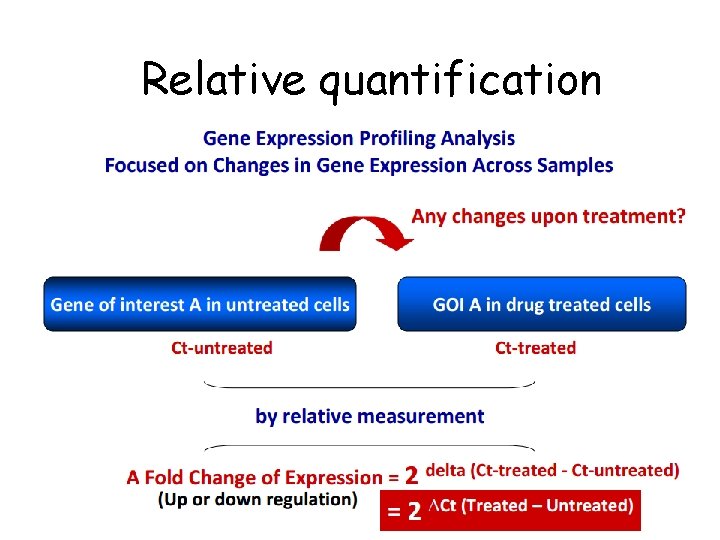
Relative quantification
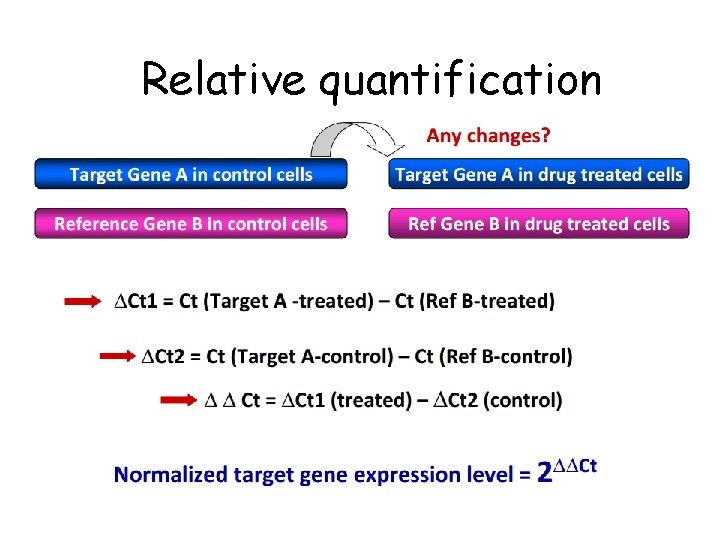
Relative quantification
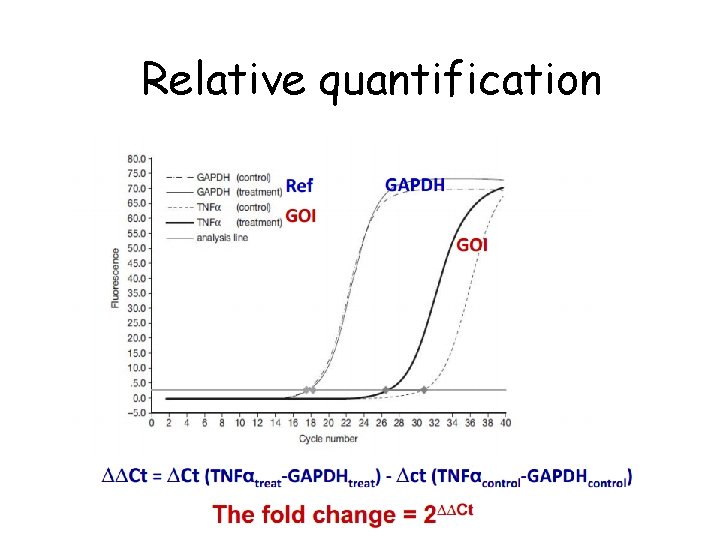
Relative quantification
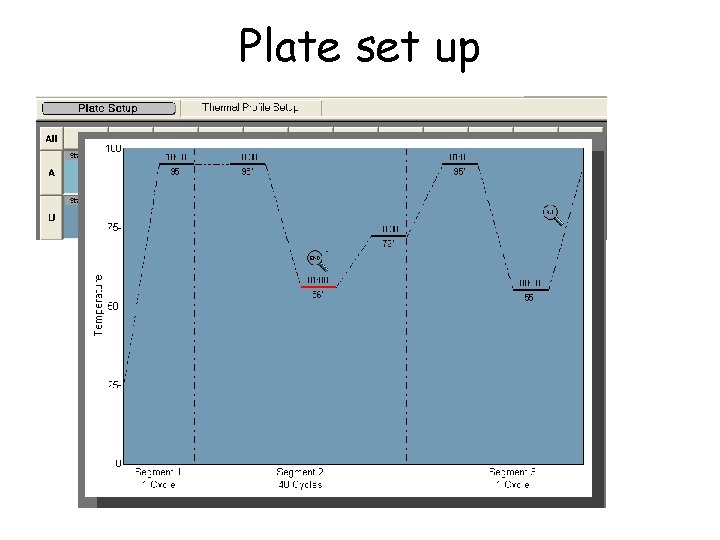
Plate set up
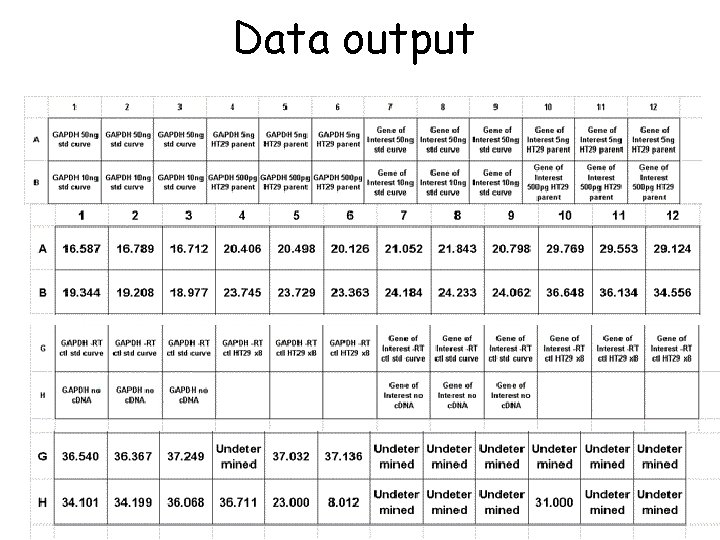
Data output

References • http: //sabiosciences. com/manuals/Intro toq. PCR. pdf • http: //relative. gene-quantification. info/ • http: //www. genomics. agilent. com/ • http: //www. protocol-online. org/ • http: //www. qiagen. com/us/products/ • https: //www. promega. com/products/
- Slides: 26