RNA isolation from monolayer cell using Trizol reagent


- Slides: 14

RNA isolation from monolayer cell using Trizol reagent Vascular Genomics Laboratory 이성운

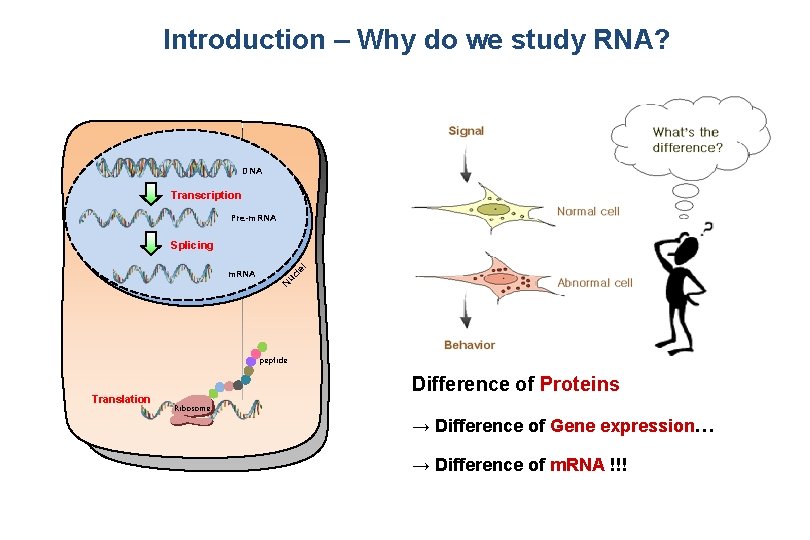
Introduction – Why do we study RNA? DNA Transcription Pre-m. RNA uc l N m. RNA ei Splicing peptide Translation Difference of Proteins Ribosome → Difference of Gene expression… → Difference of m. RNA !!!

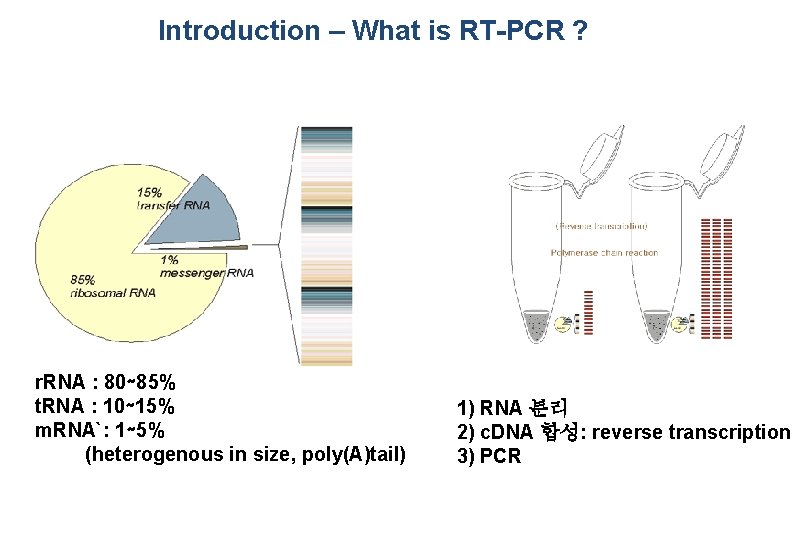
Introduction – What is RT-PCR ? r. RNA : 80∼ 85% t. RNA : 10∼ 15% m. RNA`: 1∼ 5% (heterogenous in size, poly(A)tail) 1) RNA 분리 2) c. DNA 합성: reverse transcription 3) PCR

Introduction – analysis of RNA 1. Northern hybridization - Size and amount of RNA 2. RT-PCR - Amount of RNA , c. DNA synthesis 3. RNase protection assay - Amount of RNA and mutation detection

Introduction – principle of RNA isolation 1. Controlling RNase activity - RNase inhibitors - RNase inactivation 2. Remove protein - Phenol/Chloroform 3. Separate RNA from DNA - Removal of DNA - Selection of poly(A) RNA

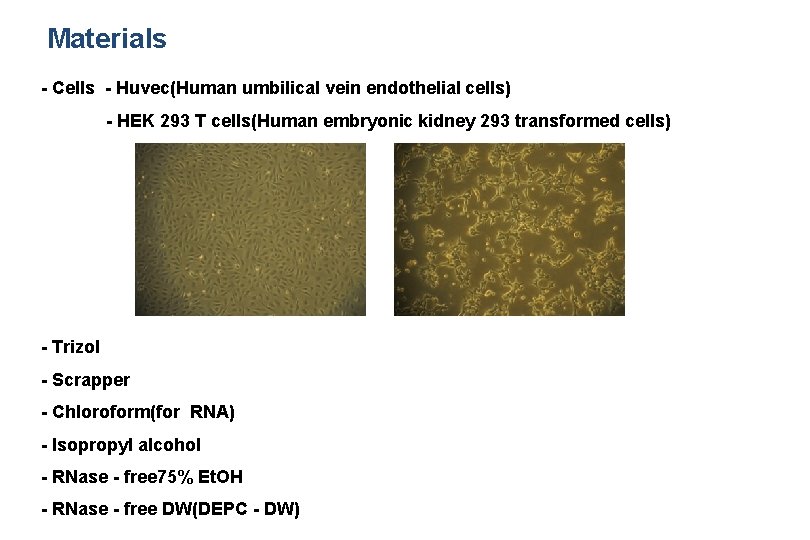
Materials - Cells - Huvec(Human umbilical vein endothelial cells) - HEK 293 T cells(Human embryonic kidney 293 transformed cells) - Trizol - Scrapper - Chloroform(for RNA) - Isopropyl alcohol - RNase - free 75% Et. OH - RNase - free DW(DEPC - DW)

Procedure 실험중 RNase에 의한 RNA degradation을 막기위해 latex glove를 반드시 착용하며 말을 자제합니다. 1. Discard the medium and add 1 ml of Trizol (per 5 -10 x 106 cells) 2. Scrape the cells with scrapper and pass the cell lysate several times through a pipette 3. Transfer the sample to 1. 5 ml tube and incubate at RT for 5 min 4. Add 0. 2 ml chloroform and shake tube vigorously by hand for 15 sec 5. Incubate at RT for 3 min 6. Centrifuge the sample at 14, 000 rpm for 15 min. 7. Transfer the aqueous phase to a fresh tube 8. Precipitate RNA by adding 0. 5 ml isopropyl alcohol and incubate samples at RT for 10 min 9. Centrifuge at 14, 000 rpm for 10 min 10. Remove the supernatant and wash the RNA pellet once with 1 ml of 75% cold Et. OH 11. Centrifuge the sample at 14, 000 rpm for 10 min. 12. At the end of the procedure, briefly dry the RNA pellet 13. Dissolve RNA in RNase-free water and incubate for 10 min at 55 to 60ºC

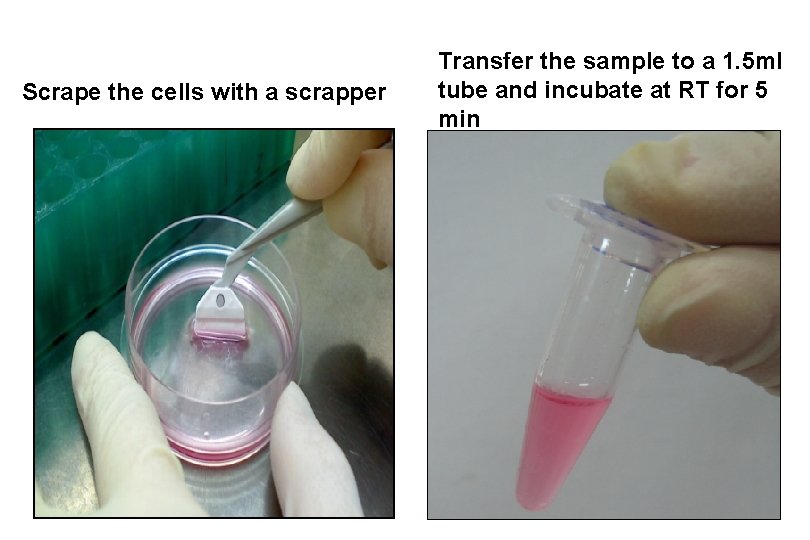
Scrape the cells with a scrapper Transfer the sample to a 1. 5 ml tube and incubate at RT for 5 min

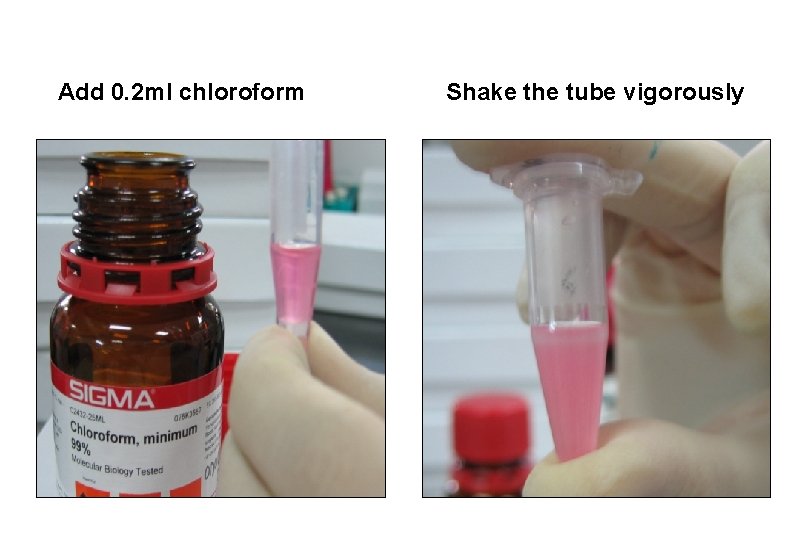
Add 0. 2 ml chloroform Shake the tube vigorously

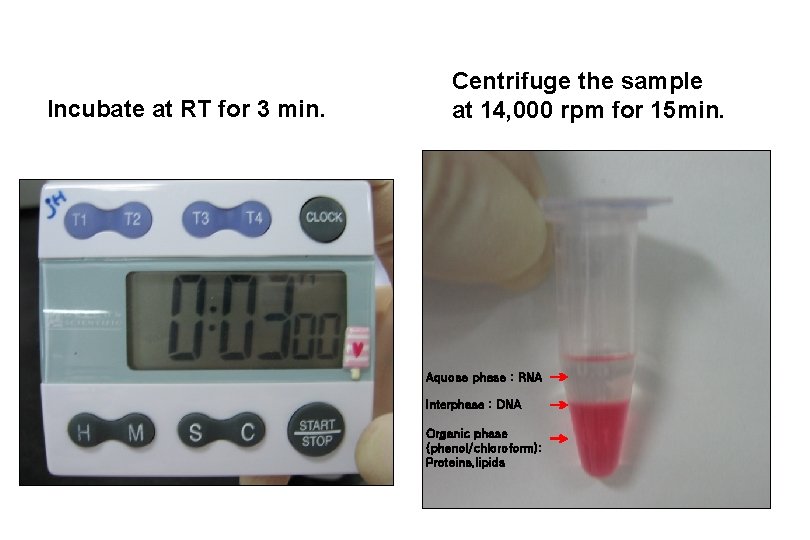
Incubate at RT for 3 min. Centrifuge the sample at 14, 000 rpm for 15 min. Aquose phase : RNA Interphase : DNA Organic phase (phenol/chloroform): Proteins, lipids

Transfer the aqueous phase to a fresh tube Add 0. 5 ml isopropyl alcohol

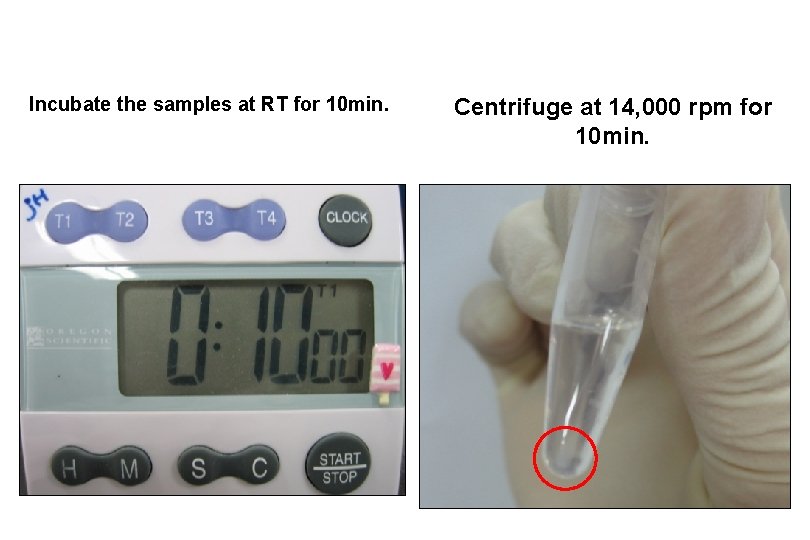
Incubate the samples at RT for 10 min. Centrifuge at 14, 000 rpm for 10 min.

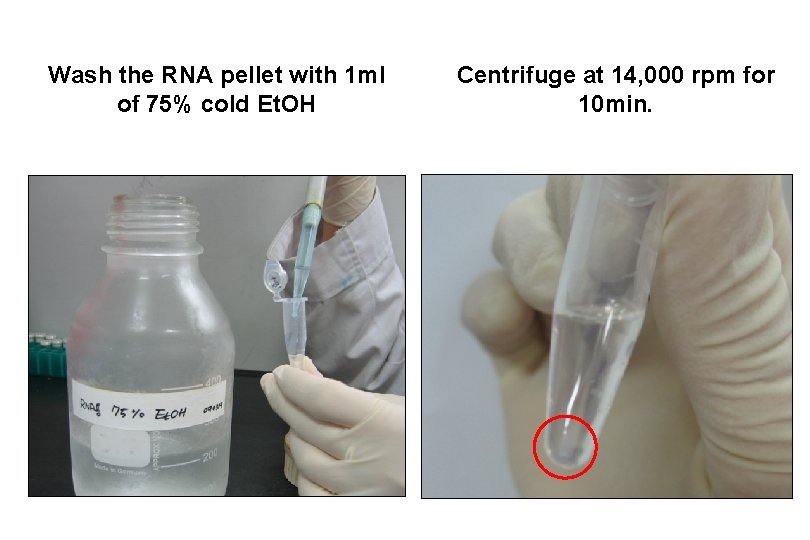
Wash the RNA pellet with 1 ml of 75% cold Et. OH Centrifuge at 14, 000 rpm for 10 min.
