RNA and Protein Synthesis How does DNA code















- Slides: 70

RNA and Protein Synthesis

How does DNA “code”? n DNA inherited by an organism ¨ Leads to specific traits by dictating synthesis of proteins n DNA directs protein synthesis/gene expression ¨ 2 stages - transcription and translation

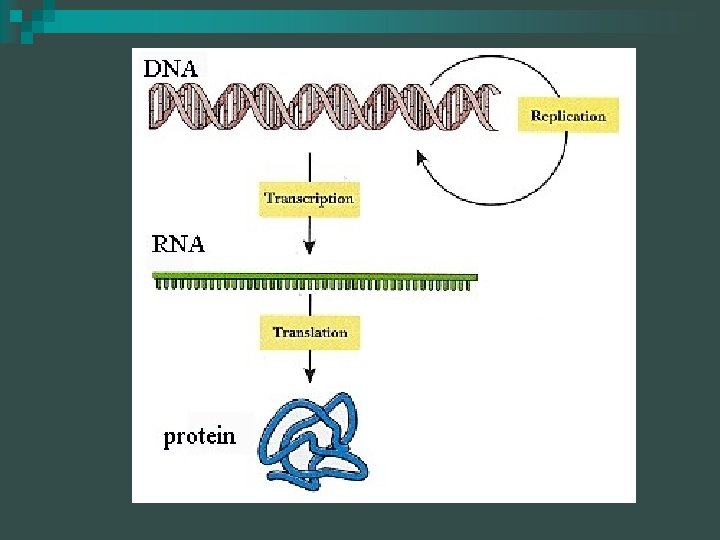
Central Dogma DNA RNA PROTEIN


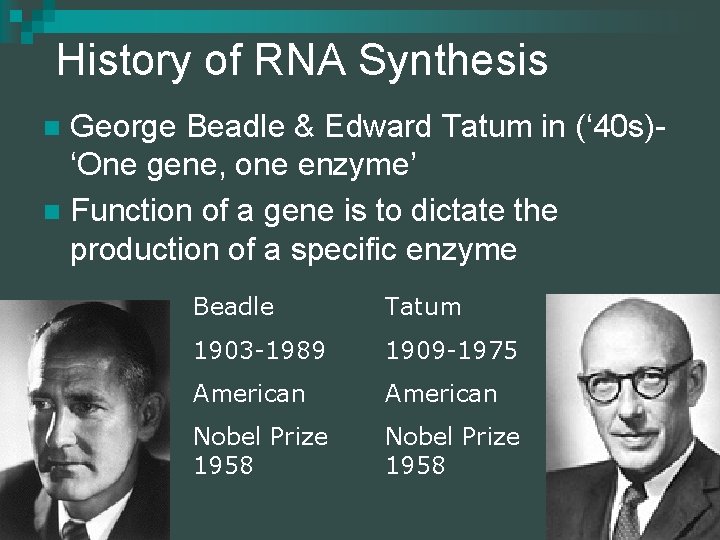
History of RNA Synthesis George Beadle & Edward Tatum in (‘ 40 s)‘One gene, one enzyme’ n Function of a gene is to dictate the production of a specific enzyme n Beadle Tatum 1903 -1989 1909 -1975 American Nobel Prize 1958

After Beadle & Tatum Some genes encode proteins that are not enzymes n One gene is responsible for one polypeptide chain, and some proteins have more than one chain n One gene, one polypeptide hypothesis n

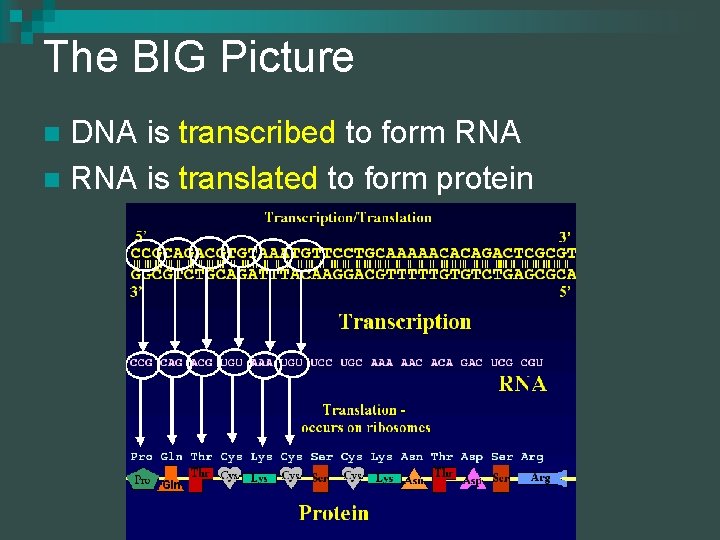
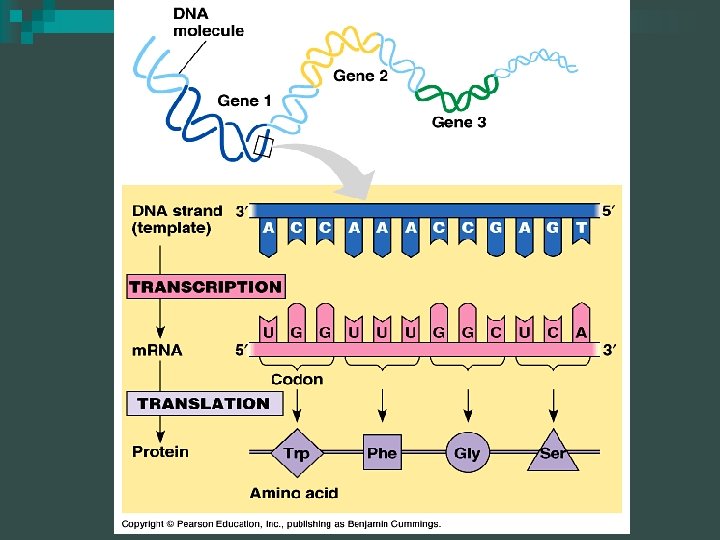
The BIG Picture DNA is transcribed to form RNA n RNA is translated to form protein n

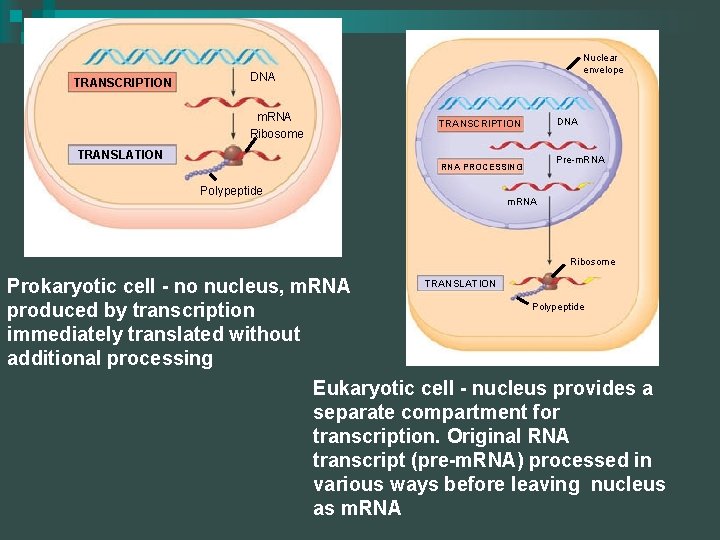
TRANSCRIPTION Nuclear envelope DNA m. RNA Ribosome TRANSCRIPTION TRANSLATION RNA PROCESSING Polypeptide DNA Pre-m. RNA Ribosome TRANSLATION Prokaryotic cell - no nucleus, m. RNA Polypeptide produced by transcription immediately translated without additional processing Eukaryotic cell - nucleus provides a separate compartment for transcription. Original RNA transcript (pre-m. RNA) processed in various ways before leaving nucleus as m. RNA

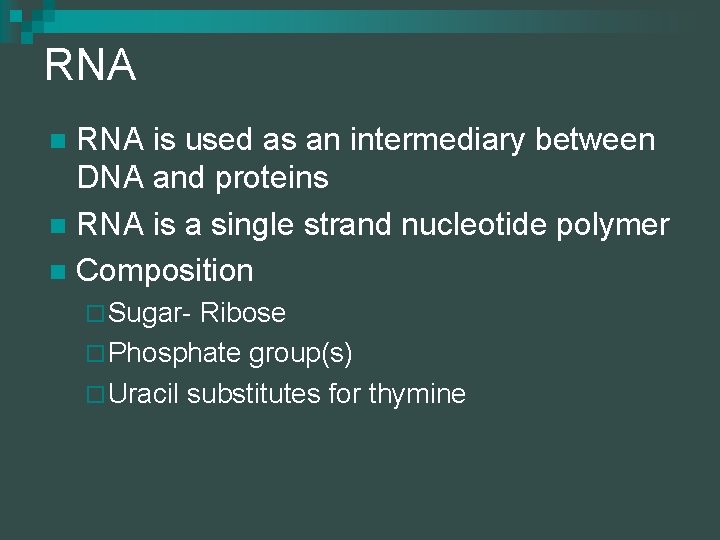
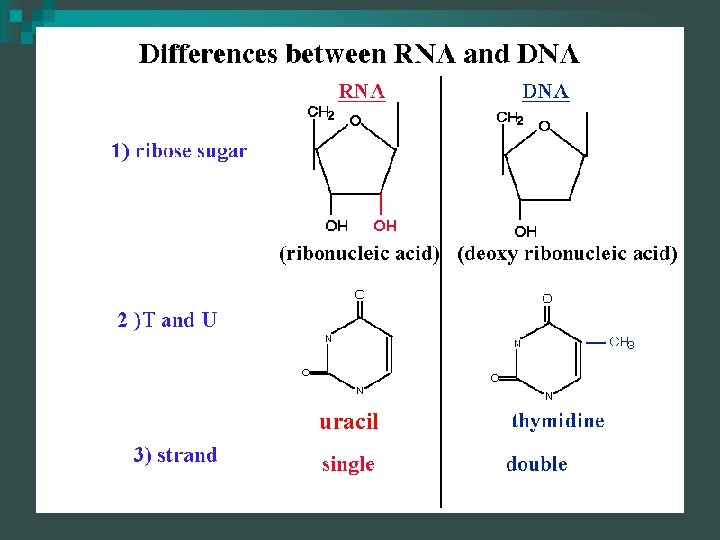
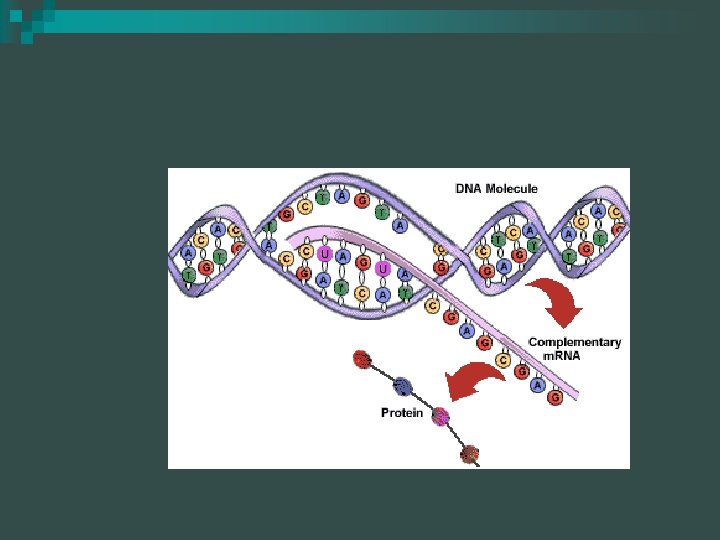
RNA is used as an intermediary between DNA and proteins n RNA is a single strand nucleotide polymer n Composition n ¨ Sugar- Ribose ¨ Phosphate group(s) ¨ Uracil substitutes for thymine

uracil

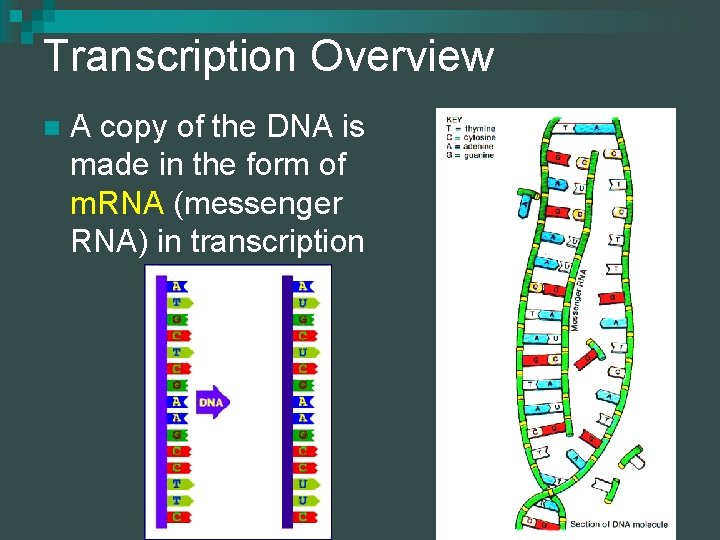
Transcription Overview n A copy of the DNA is made in the form of m. RNA (messenger RNA) in transcription


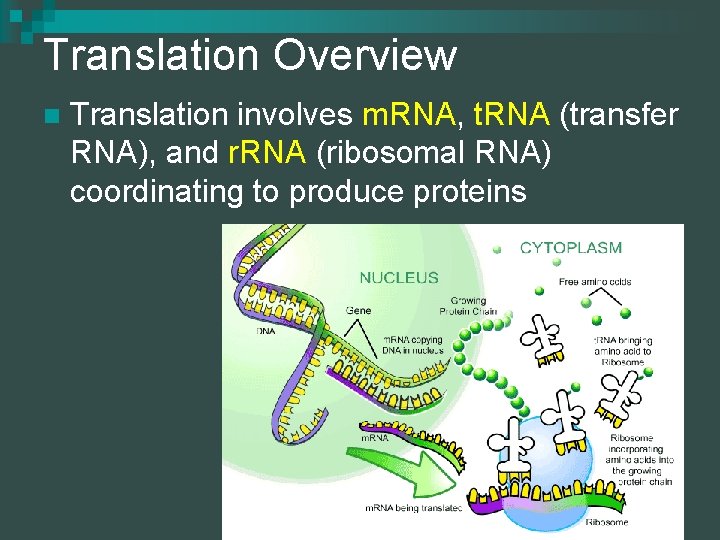
Translation Overview n Translation involves m. RNA, t. RNA (transfer RNA), and r. RNA (ribosomal RNA) coordinating to produce proteins

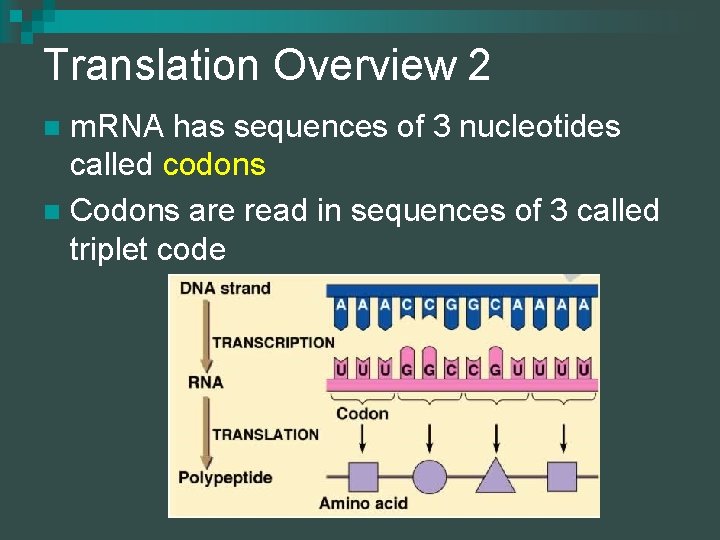
Translation Overview 2 m. RNA has sequences of 3 nucleotides called codons n Codons are read in sequences of 3 called triplet code n


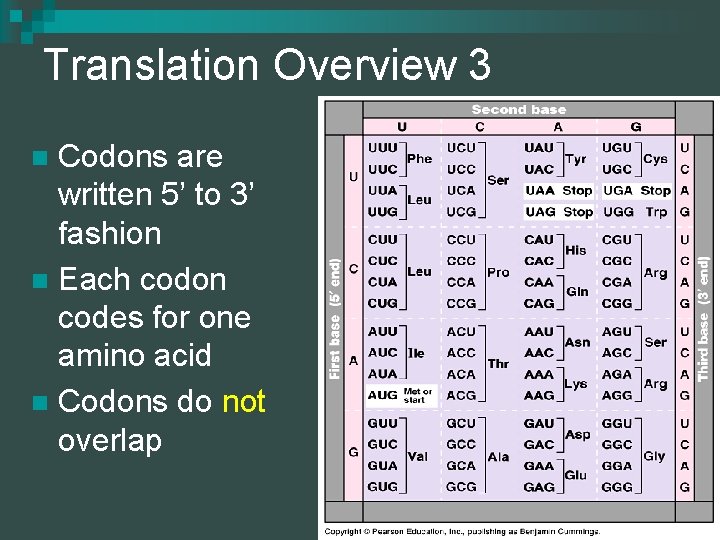
Translation Overview 3 Codons are written 5’ to 3’ fashion n Each codon codes for one amino acid n Codons do not overlap n

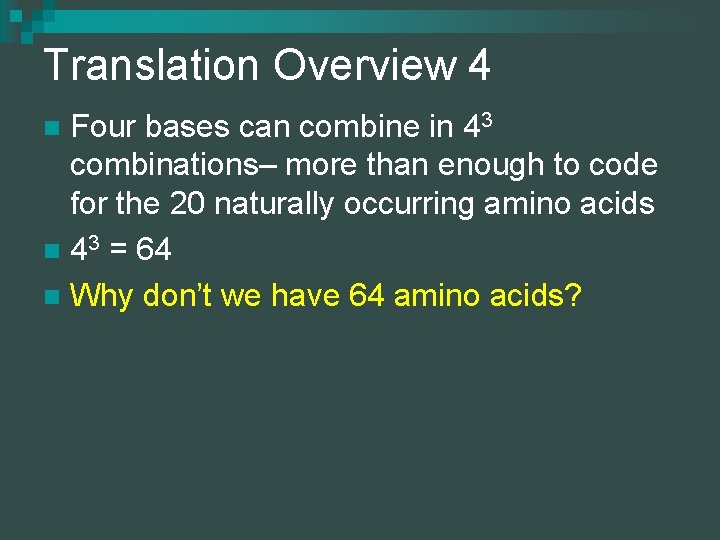
Translation Overview 4 Four bases can combine in 43 combinations– more than enough to code for the 20 naturally occurring amino acids n 43 = 64 n Why don’t we have 64 amino acids? n

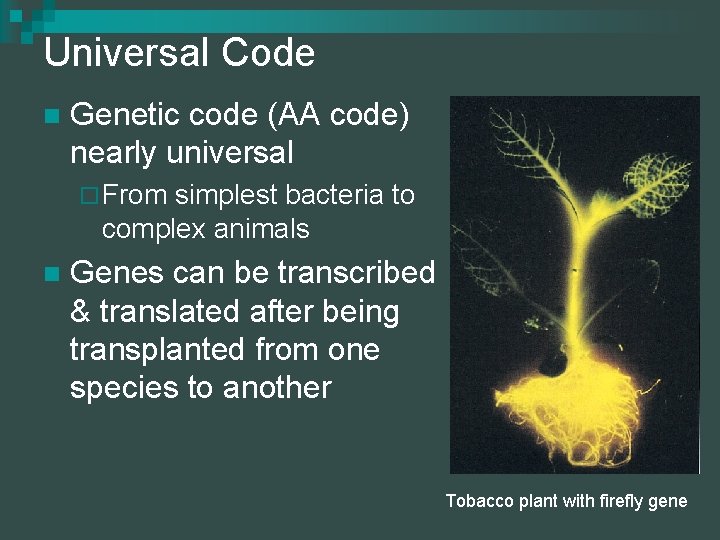
Universal Code n Genetic code (AA code) nearly universal ¨ From simplest bacteria to complex animals n Genes can be transcribed & translated after being transplanted from one species to another Tobacco plant with firefly gene

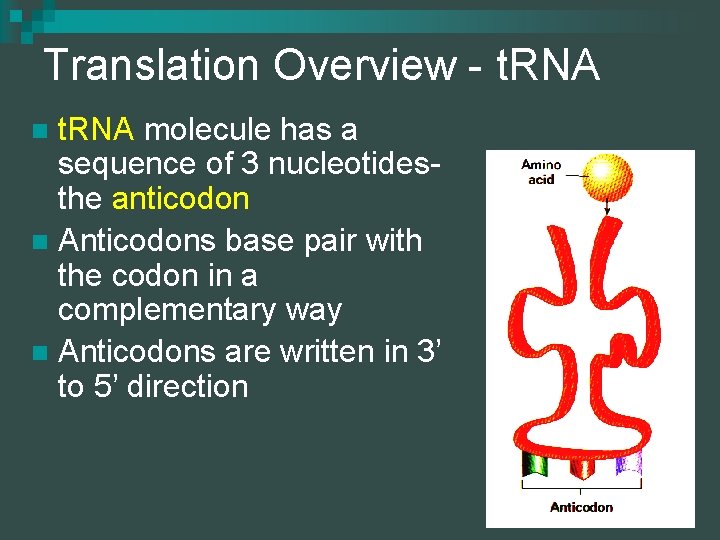
Translation Overview - t. RNA molecule has a sequence of 3 nucleotidesthe anticodon n Anticodons base pair with the codon in a complementary way n Anticodons are written in 3’ to 5’ direction n


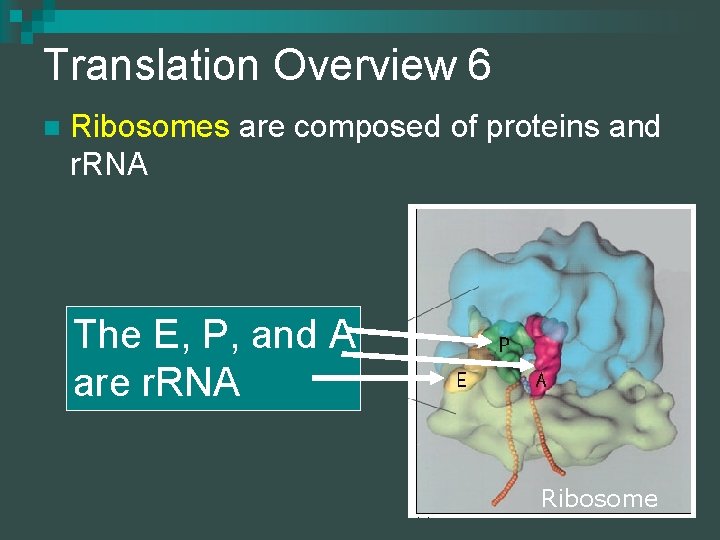
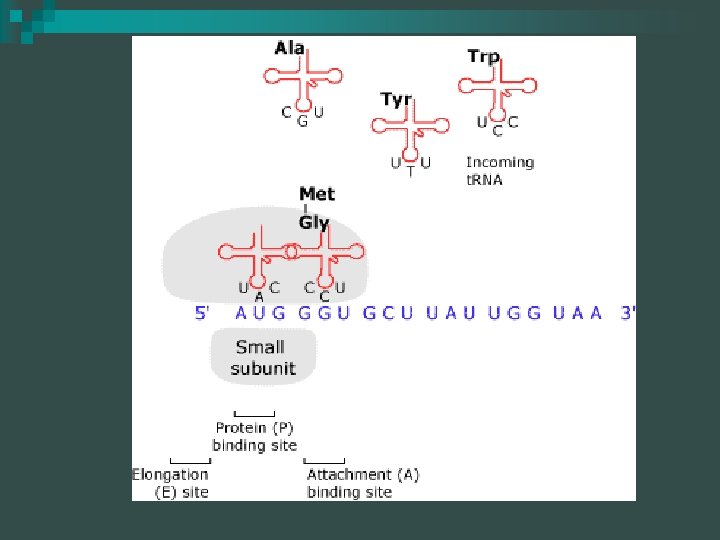
Translation Overview 6 n Ribosomes are composed of proteins and r. RNA The E, P, and A are r. RNA Ribosome

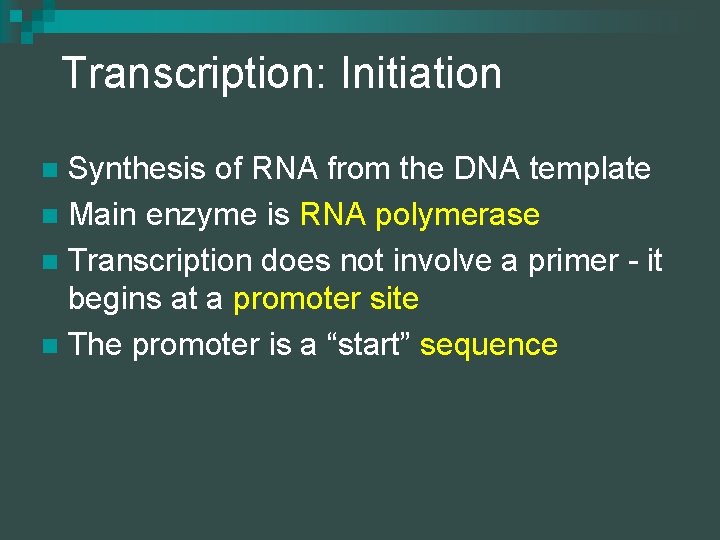
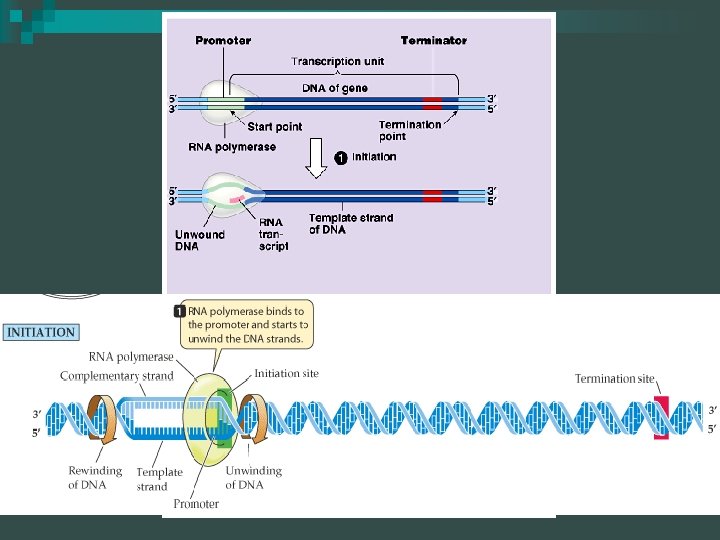
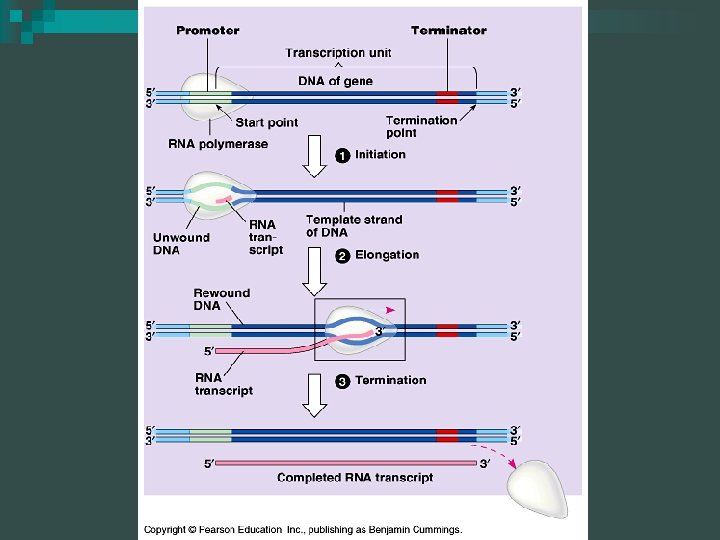
Transcription: Initiation Synthesis of RNA from the DNA template n Main enzyme is RNA polymerase n Transcription does not involve a primer - it begins at a promoter site n The promoter is a “start” sequence n


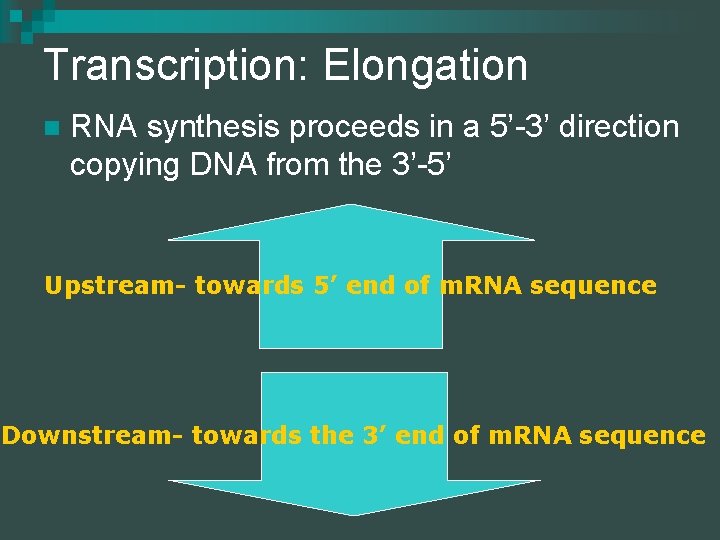
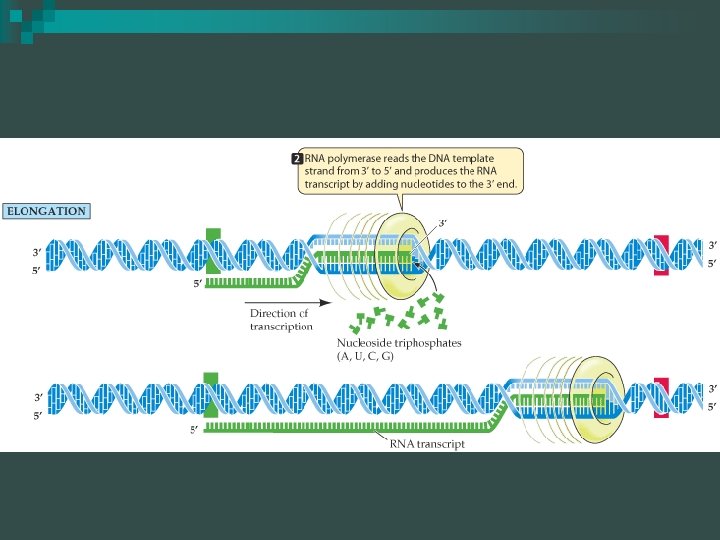
Transcription: Elongation n RNA synthesis proceeds in a 5’-3’ direction copying DNA from the 3’-5’ Upstream- towards 5’ end of m. RNA sequence Downstream- towards the 3’ end of m. RNA sequence

Transcription: Elongation RNA polymerase moves along the DNA n Untwists the double helix, exposing about 10 to 20 DNA bases at a time for pairing with RNA nucleotides n


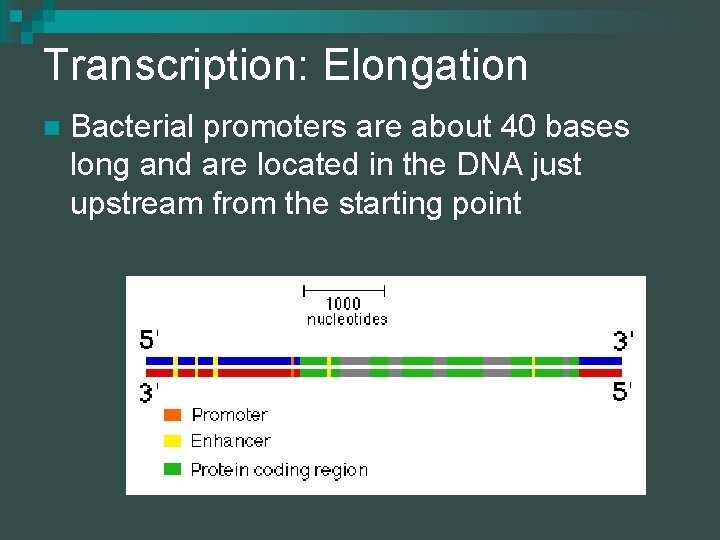
Transcription: Elongation n Bacterial promoters are about 40 bases long and are located in the DNA just upstream from the starting point

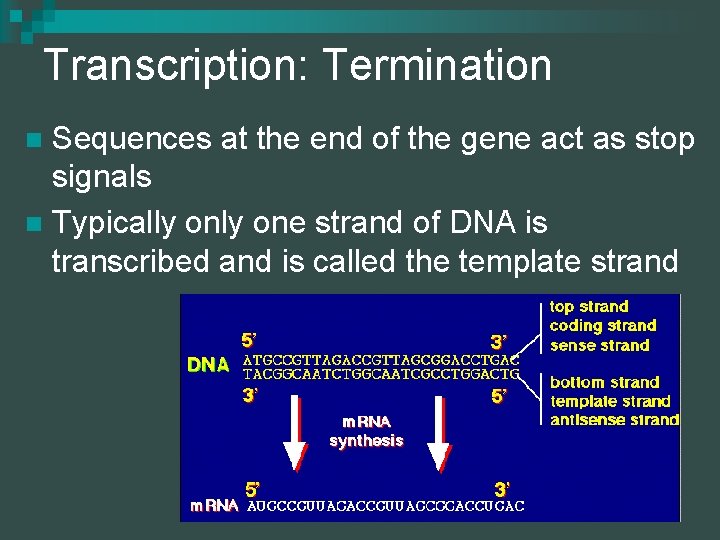
Transcription: Termination Sequences at the end of the gene act as stop signals n Typically one strand of DNA is transcribed and is called the template strand n


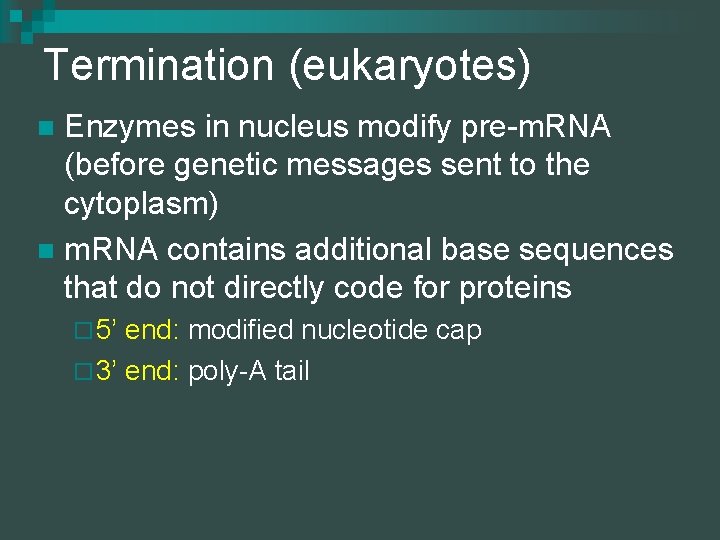
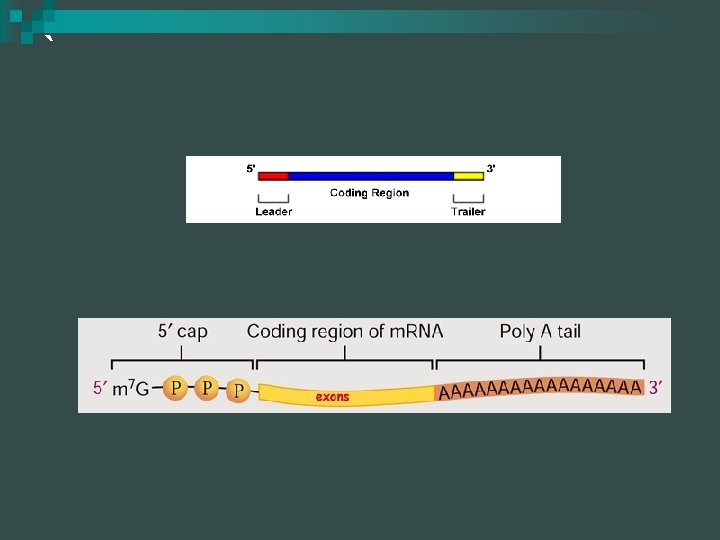
Termination (eukaryotes) Enzymes in nucleus modify pre-m. RNA (before genetic messages sent to the cytoplasm) n m. RNA contains additional base sequences that do not directly code for proteins n ¨ 5’ end: modified nucleotide cap ¨ 3’ end: poly-A tail


5’ Cap & Poly A Tail n 5’ Cap ¨ Protect m. RNA from hydrolytic enzymes ¨ Functions as an “attach here” signal for ribosomes n Poly A Tail ¨ 50 – 250 nucleotides ¨ Same function as 5’ cap ¨ Faciliates export from nucleus

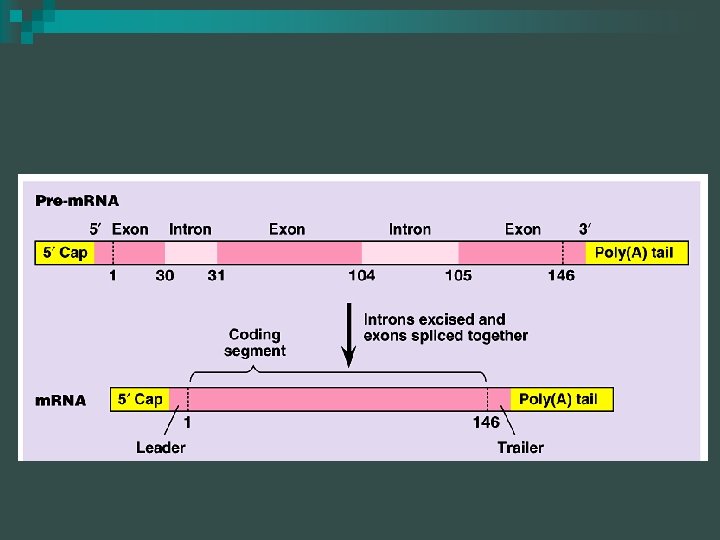
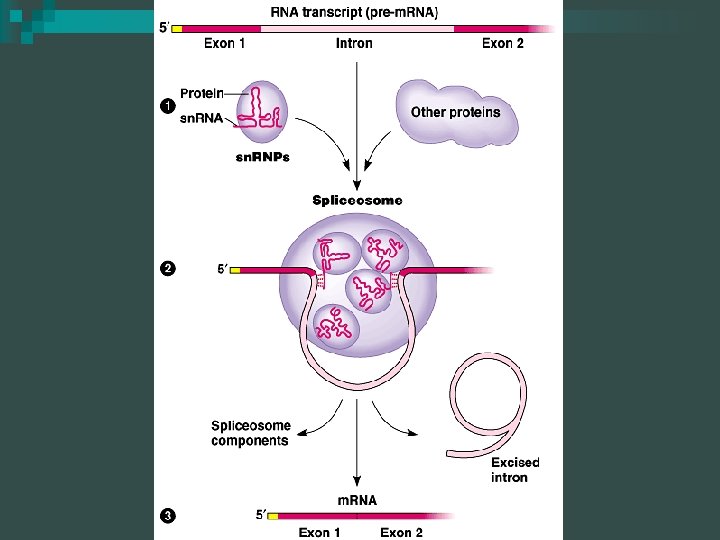
RNA Modification n RNA splicing ¨ Removes introns and joins exons ¨ Introns – noncoding regions of DNA ¨ Exons – coding portions of DNA ¨ Introns stay “in” the nucleus, exons “exit” the nucleus


Spliceosome n Splicing accomplished by a spliceosome ¨ Consists of a variety of proteins and several small nuclear ribonucleoproteins (sn. RNPs) ¨ Each sn. RNP has several protein molecules and a small nuclear RNA molecule (sn. RNA) ¨ Each is about 150 nucleotides long


Translation 1 During translation, the nucleic acid message is decoded n An amino acid is attached to t. RNA before becoming incorporated into a polypeptide n ¨ To form a polypeptide chain, the amino and carboxyl groups of amino acids are joined



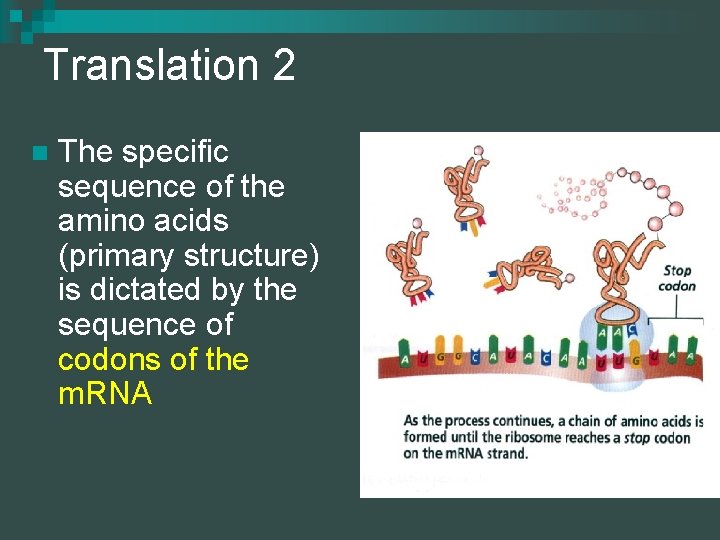
Translation 2 n The specific sequence of the amino acids (primary structure) is dictated by the sequence of codons of the m. RNA

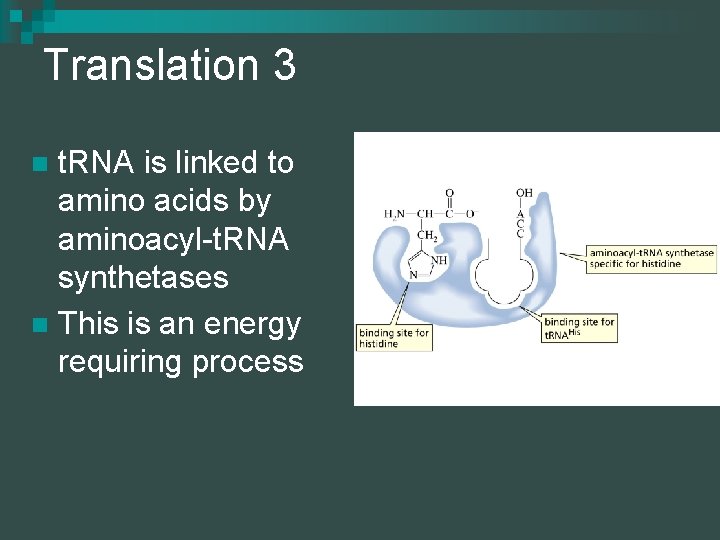
Translation 3 t. RNA is linked to amino acids by aminoacyl-t. RNA synthetases n This is an energy requiring process n

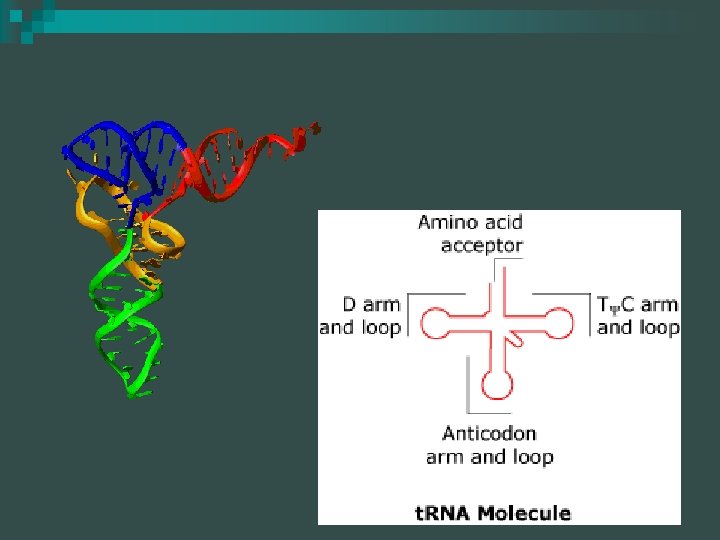
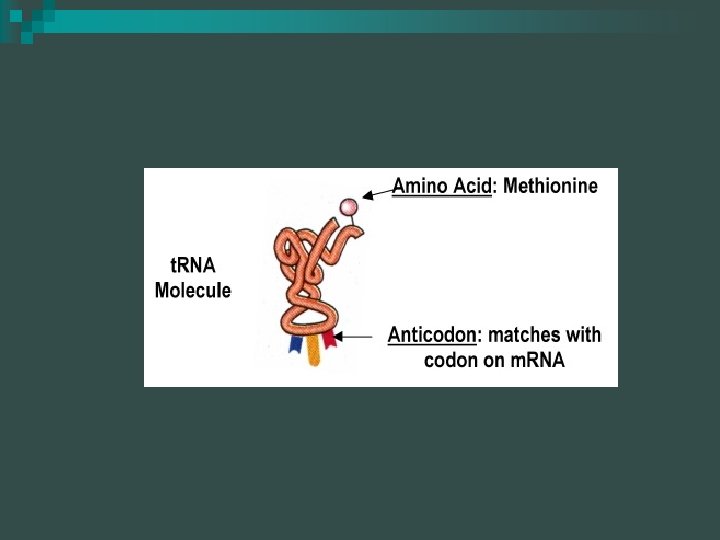
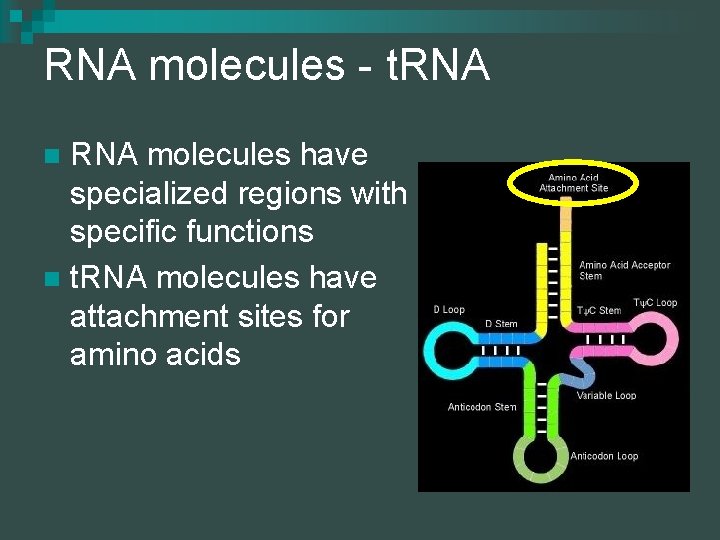
RNA molecules - t. RNA molecules have specialized regions with specific functions n t. RNA molecules have attachment sites for amino acids n

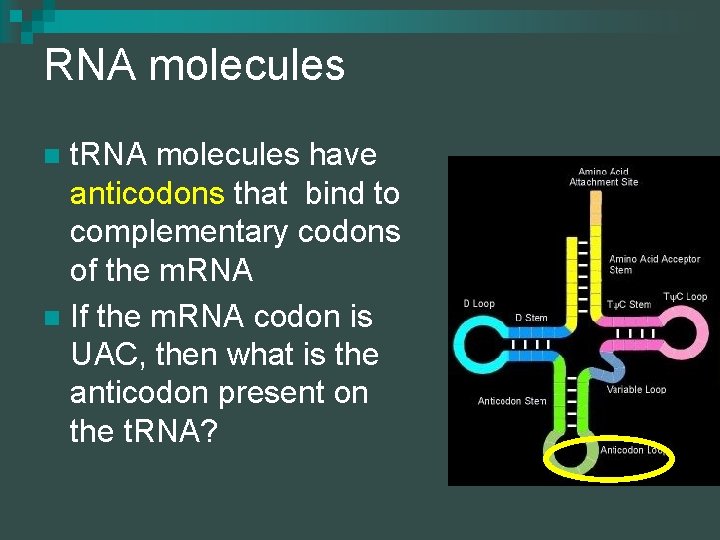
RNA molecules t. RNA molecules have anticodons that bind to complementary codons of the m. RNA n If the m. RNA codon is UAC, then what is the anticodon present on the t. RNA? n

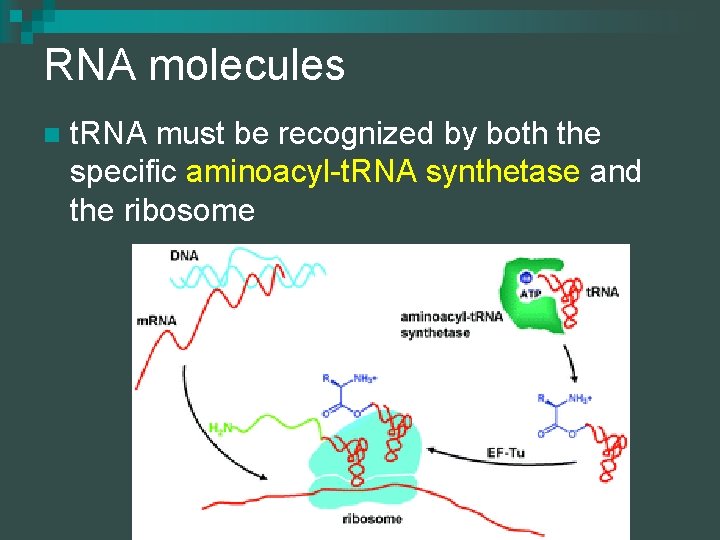
RNA molecules n t. RNA must be recognized by both the specific aminoacyl-t. RNA synthetase and the ribosome

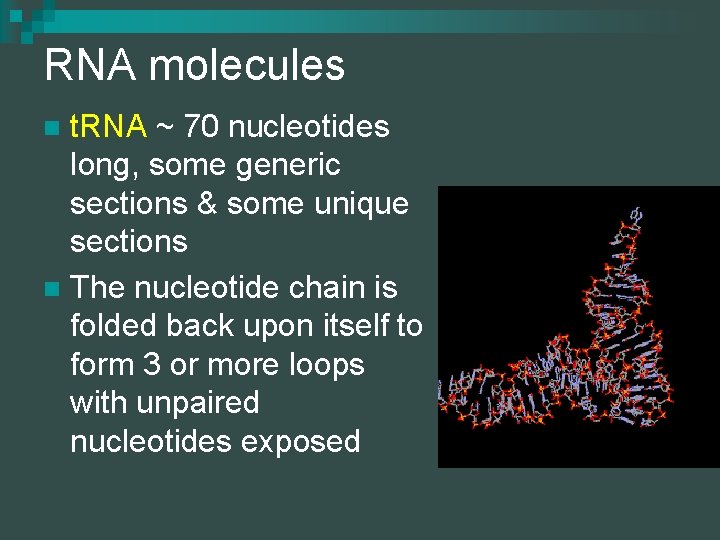
RNA molecules t. RNA ~ 70 nucleotides long, some generic sections & some unique sections n The nucleotide chain is folded back upon itself to form 3 or more loops with unpaired nucleotides exposed n

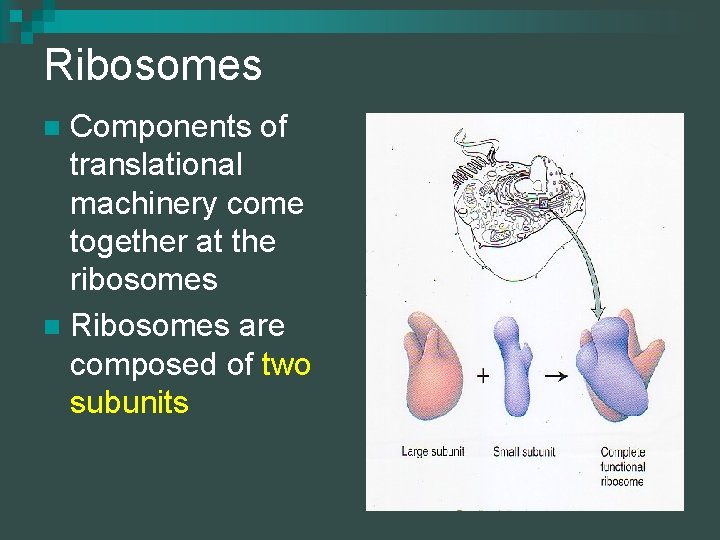
Ribosomes Components of translational machinery come together at the ribosomes n Ribosomes are composed of two subunits n

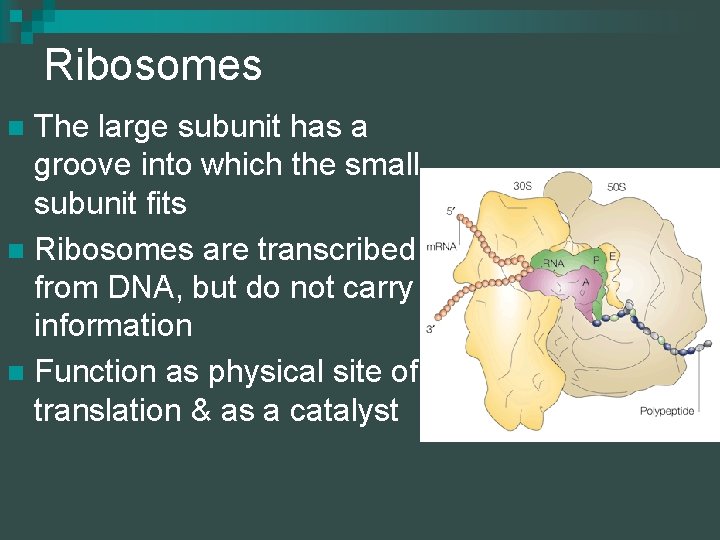
Ribosomes The large subunit has a groove into which the small subunit fits n Ribosomes are transcribed from DNA, but do not carry information n Function as physical site of translation & as a catalyst n

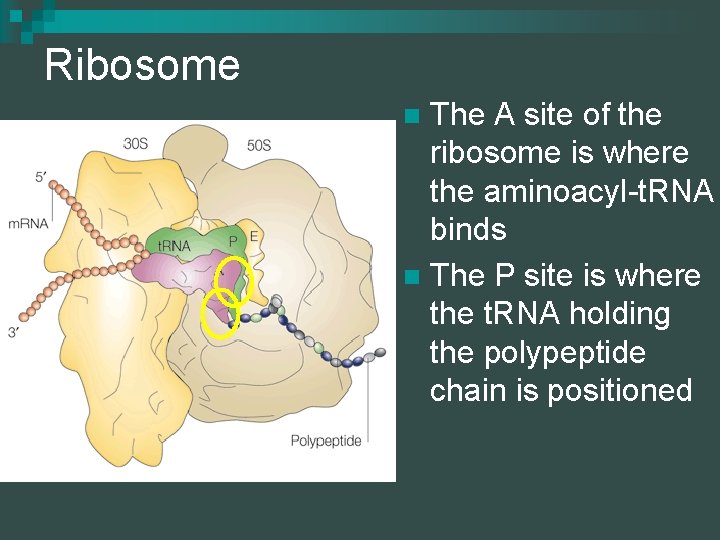
Ribosome The A site of the ribosome is where the aminoacyl-t. RNA binds n The P site is where the t. RNA holding the polypeptide chain is positioned n

Translation Steps Inititiation n Elongation n Termination n

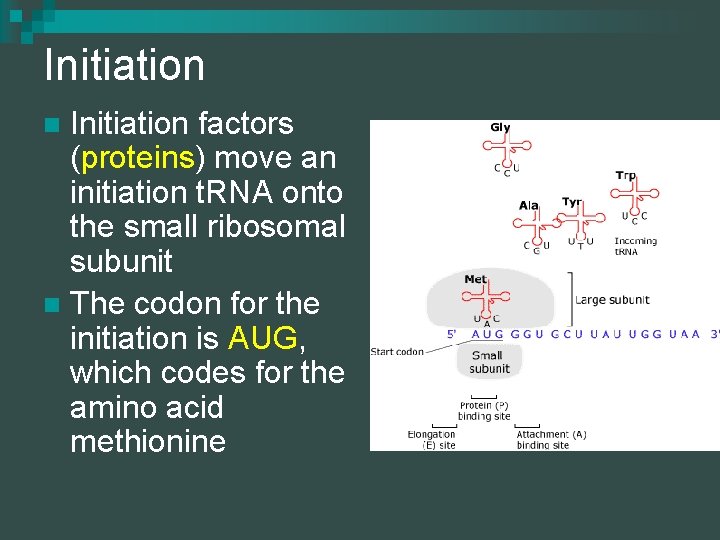
Initiation factors (proteins) move an initiation t. RNA onto the small ribosomal subunit n The codon for the initiation is AUG, which codes for the amino acid methionine n

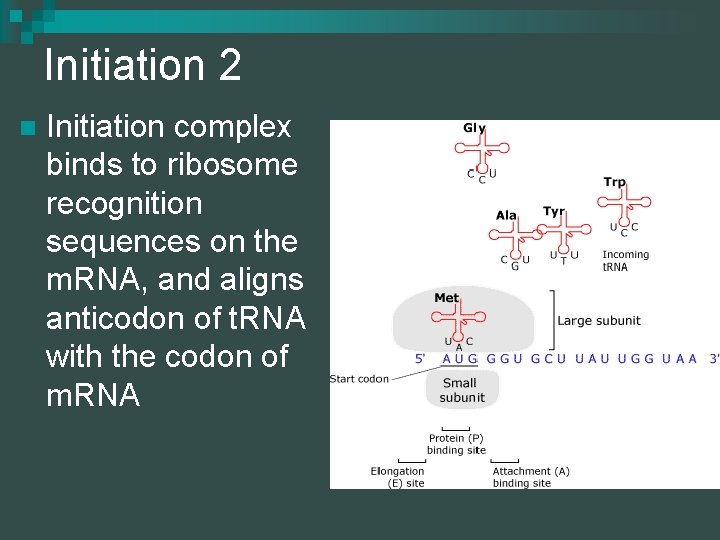
Initiation 2 n Initiation complex binds to ribosome recognition sequences on the m. RNA, and aligns anticodon of t. RNA with the codon of m. RNA

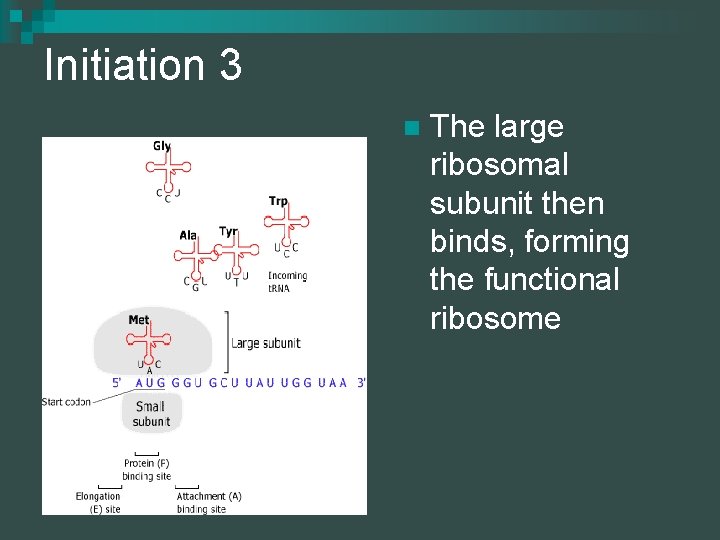
Initiation 3 n The large ribosomal subunit then binds, forming the functional ribosome

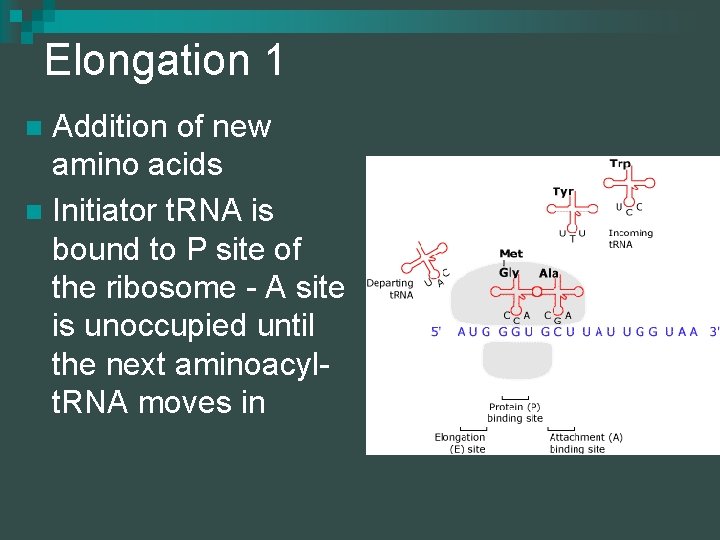
Elongation 1 Addition of new amino acids n Initiator t. RNA is bound to P site of the ribosome - A site is unoccupied until the next aminoacylt. RNA moves in n

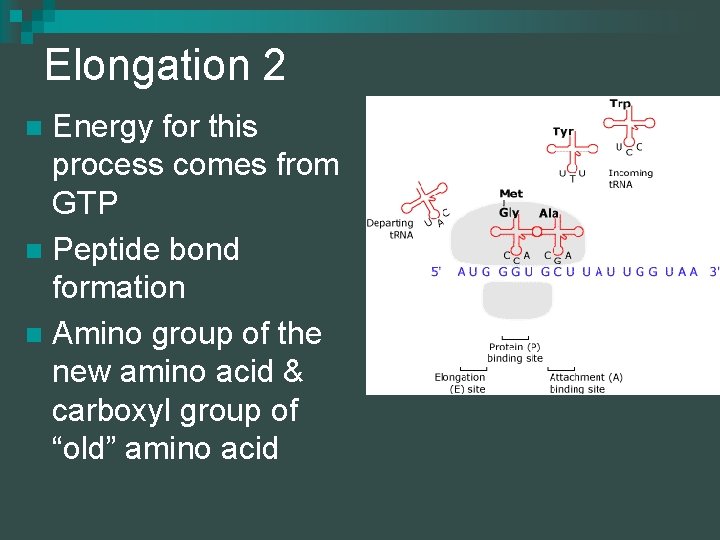
Elongation 2 Energy for this process comes from GTP n Peptide bond formation n Amino group of the new amino acid & carboxyl group of “old” amino acid n

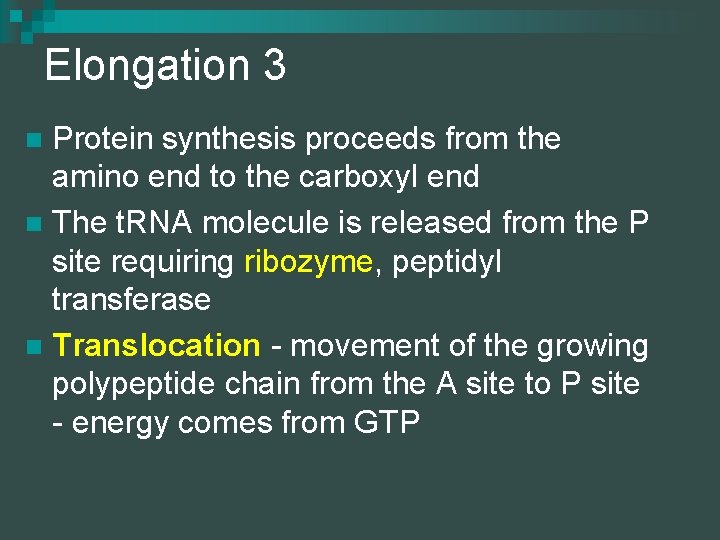
Elongation 3 Protein synthesis proceeds from the amino end to the carboxyl end n The t. RNA molecule is released from the P site requiring ribozyme, peptidyl transferase n Translocation - movement of the growing polypeptide chain from the A site to P site - energy comes from GTP n

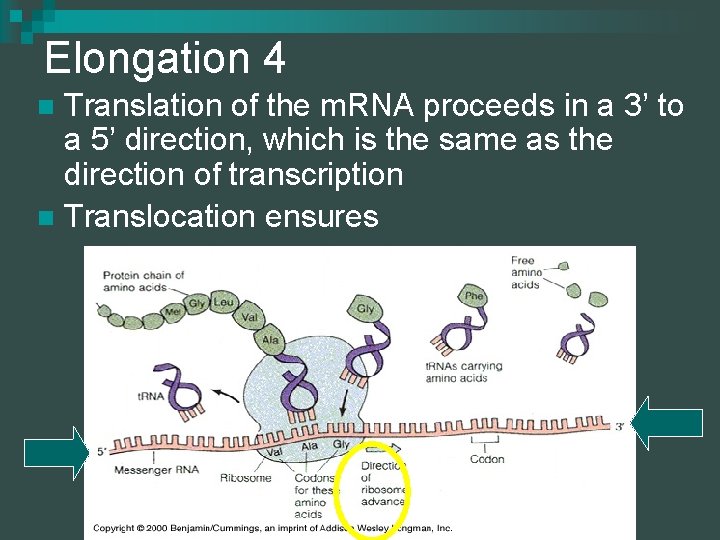
Elongation 4 Translation of the m. RNA proceeds in a 3’ to a 5’ direction, which is the same as the direction of transcription n Translocation ensures n

Termination Occurs when the m. RNA presents the codons UAA, UGA, or UAG; n No complementary t. RNA n Release factors recognize codons n The ribosome dissociates into the two subunits n

Translation Tutorial n http: //telstar. ote. cmu. edu/Hughes Archive/tutorial/polypeptide/tutorial. swf

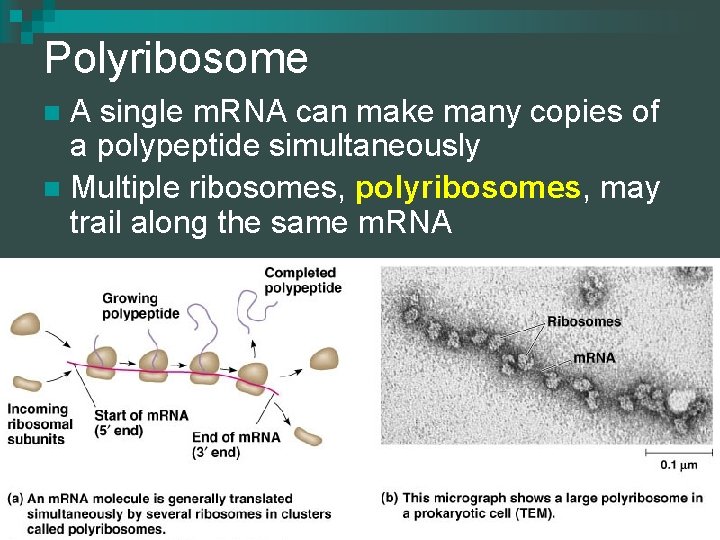
Polyribosome A single m. RNA can make many copies of a polypeptide simultaneously n Multiple ribosomes, polyribosomes, may trail along the same m. RNA n

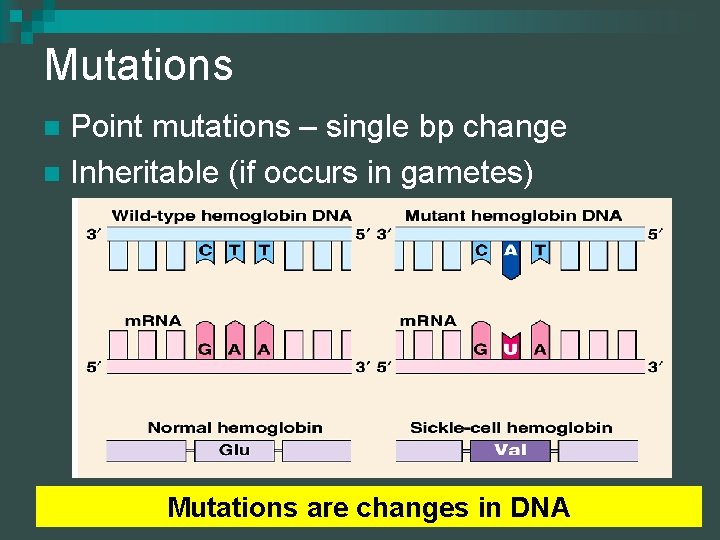
Mutations Point mutations – single bp change n Inheritable (if occurs in gametes) n Mutations are changes in DNA

Point mutation n Base pair substitution ¨ Replacement of a pair of complementary nucleotides with another nucleotide pair Some have little/no impact n Silent mutation n AA change n ¨ Similar ¨ Nonessential

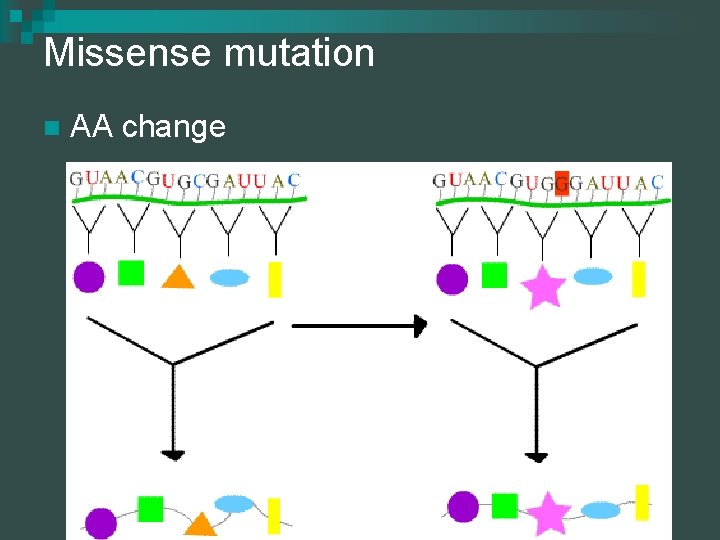
Missense mutation n AA change

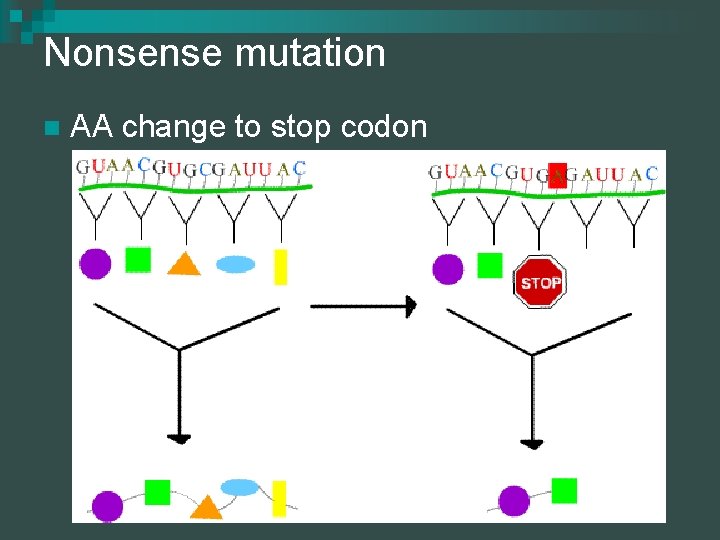
Nonsense mutation n AA change to stop codon

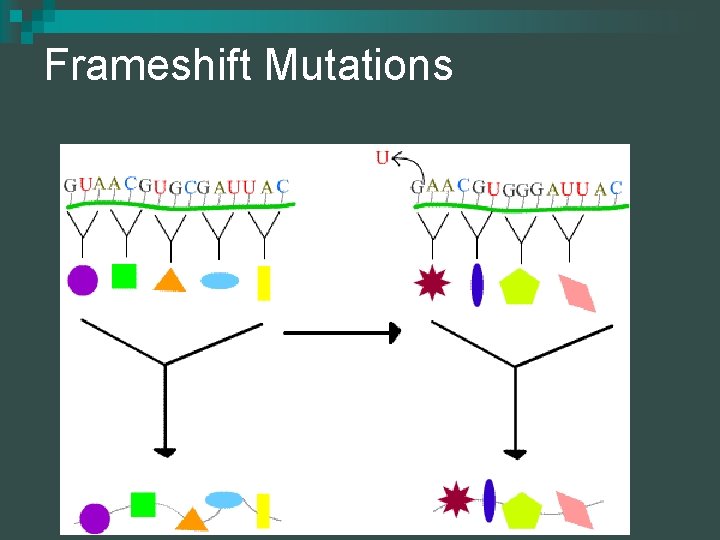
Mutations Frameshift mutations- involve the insertion or deletion of a base n They result in an entirely different sequence of amino acids because they change the ________ n Mutations are changes in DNA

Frameshift Mutations

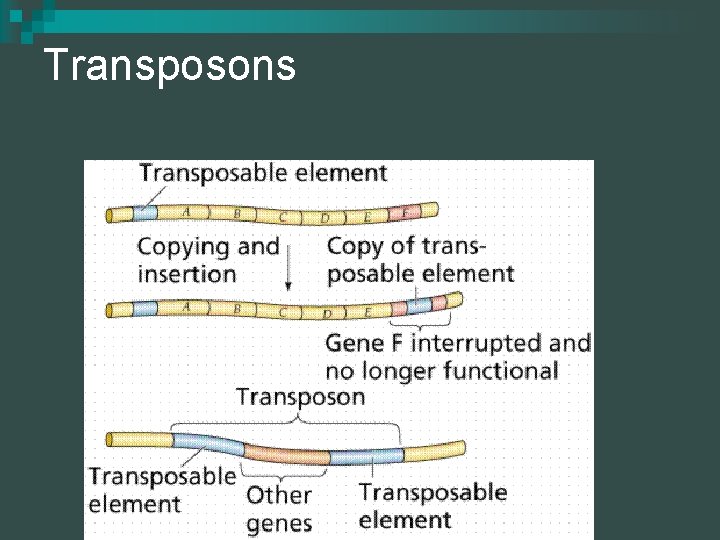
Mutations n Transposons - movable sequences of DNA that may move into another area of DNA ¨ May disrupt genes, but may inactivate others ¨ Transposons have some similarities to retroviruses Mutations are changes in DNA

Transposons

Barbara Mc. Clintock (1902 -1992) n n Nobel Prize 1983 Cornell University Discovered the transposons “jumping genes” Studied maize

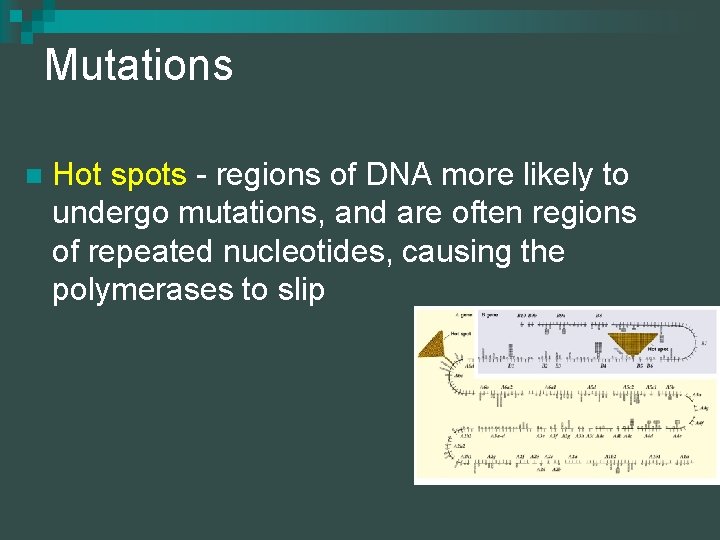
Mutations n Hot spots - regions of DNA more likely to undergo mutations, and are often regions of repeated nucleotides, causing the polymerases to slip

Mutagens n Agents that cause mutations ¨ Including ionizing radiation ¨ Mutations in somatic cells are not passed on to the next generation ¨ Some mutagens are also carcinogens