Ribosymes RNA Enzymes OVERVIEW STRUCTURE What Do They

Ribosymes: RNA Enzymes OVERVIEW & STRUCTURE

What Do They Do? v Ribonucleic acid enzymes v RNA molecules that act as chemical catalysts v Many work by cutting, patching, and moving RNA around. v GGCCUACCGG v Work through base pair interactions. Will end up foiling on it’s own.

5 Classes of Ribozymes: Hepatitis Delta Virus Hammerhead Tetrahymena Group I Intron Rnase P Hairpin

The Discovery of the Ribozyme In 1989, Thomas R. Cech and Sidney Altman won the Nobel Prize in chemistry for their "discovery of catalytic properties of RNA. " Before the discovery of Ribozymes, enzymes were the only know biological catalysts.

The Tetrahymena Ribozyme The 3′-terminal guanosine (ωG) serves as the attacking group for RNA cleavage v Catalyzes phosphoryl transfer between guanosine and a substrate RNA. v. Metal ion present in the active site.

Metal Ion at the Active Site Metal ions, particularly divalent cations, are essential for the function of most larger ribozymes v. Required for structural stability and formation of the active site. v. Plays direct role in catalysis. v. The oxygen atoms on the three phosphates close to the magnesium ion are indicated as red.

The A-rich Bulge Region has high affinity for divalent cations. Folds first upon addition of magnesium.

GNRA Tetraloop Many of the structural motifs important for domain folding involve adenosine-rich sequences. v A GAAA tetraloop in the p 5 b domain specifically interacts with an 11 nucleotide region in the p 6 a domain that contains a internal loop.

Adenosine Platform v The A-A platform motif consists of adjacent adenosines in a helical strand that form a pseudo base pair within the helix. v Driving force of platform is base stacking. Wobble base pairs indicated in red.

The Hammerhead Ribozyme v Consists of three helices that interact at a highly conserved catalytic core of 11 necleotides. v Supports cleavage at high concentrations of Mg+

Minimal Hammerhead Ribozyme v. Wish bone shape v 16 -nt enzyme stand (red) v 25 -nt substrate strand (yellow) v. C 17 is the cleavage-site base

The Sequence and the Fold Full length Hammerhead v Tertiary contacts distant from the active site forms between Stem II and unpaired region of Stem I. v The Stem I—Stem II interaction accelerates the ribozyme reaction 1000 fold. Secondary Structure Tertiary Structure

The Active Site v. The A-9 and Scissile phosphates are said to form a single metal binding site v. G-12 positioned for general base catalysis v. G-8 may assist as a general acid

Highly Conserved Sequences v. Three stems are all A-form helices. v. Two G-A base pairs and one A-U base pair. v. CUGA sequence near cleavage site.

Divalent Metal Ions v These ions have both structural and catalytic roles. v Two-stage ion dependent folding process.

Function • Ribozymes • The ribosome is a ribozyme • Translates RNA into proteins • Responsible for ribosome structure • Ability to position t. RNAs on m. RNA • 3 o structure • Folds to serve as an enzyme • Targets RNA • Metal ions – structural stability and formation at active site • RNA segments • Form covalent bonds • Act as molecular scissors • Catalytic activity • Cleavage • Splicing • Ligation • RNA chains • Main Role of Ribozyme: stabilize the RNA core to catalyze protein synthesis

Principal RNAs produced in Cells Type of RNA Function m. RNA (messenger RNA) code for proteins r. RNA (ribosomal RNA) form the basic structure of the ribosome and catalyze protein synthesis t. RNA (transfer RNA) central to protein synthesis sn. RNA (small nuclear RNA) Function in a variety of nuclear processes →splicing of prem. RNA sno. RNA (small nucleolar RNA) Used to process and chemically modify r. RNA sca. RNA (small cajal RNA) Used to modify sno. RNA and sn. RNA mi. RNA (micro. RNA) Regulate gene expression typically by blocking translation of selective m. RNA si. RNA (small interfering RNA) Turn off gene expression by directing degradation of selective m. RNA and the establishment of compact chromatin structures Other noncoding RNA Function in diverse cell processes → telomere synthesis, X-chromosome inactivation, and the transport of proteins into the ER Molecular Biology of the Cell (© Garland Science 2008)

Cleaving, Ligating, Splicing Activity Ribozymes Peptide bond formation in protein synthesis r. RNA Cleavage, Litagation self-splicing RNA; RNase P; in vitro selected RNA DNA Cleavage self-splicing RNA splicing self-splicing RNA polymerization in vitro selected RNA Range of catalytic activities

Cleaving RNA Molecular Biology of the Cell (© Garland Science 2008)

Self-Cleaving Hydrolysis of a ribozyme’s own phosphodiester bond Hepatitis Delta Virus (HDV)

Self-Ligating Joining together with a bond The ligation reaction forms a 2’, 5’phosphodiester

Self-Splicing Ribozyme built into their own intron and is degraded Introns and RNase P
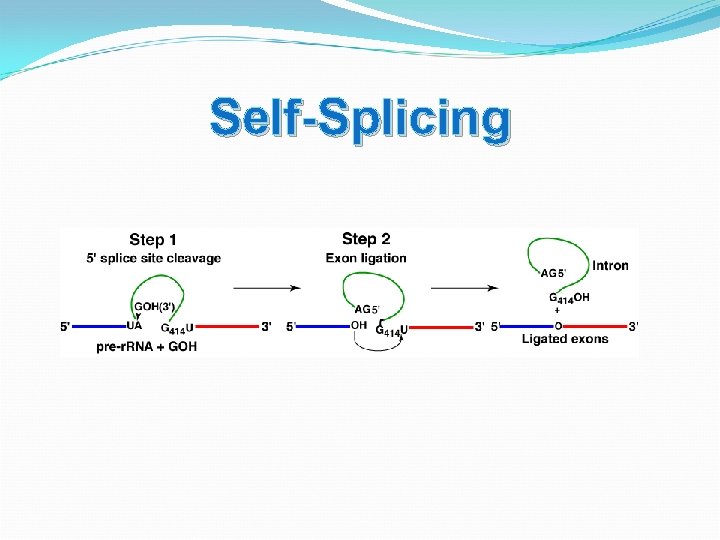
Self-Splicing

Hepatitis Delta Virus Noncoding RNA - function in diverse cell processes 1, 700 nucleotides 70% self-complementarity Essential in viral replication the only catalytic RNA known to be required for the viability of a human pathogen Self-cleaving active in vitro in the absence of any proteins

HDV Purpose - cleave rolling circle replication products into genomelength units Fastest naturally occurring catalyst folded into a double pseudoknot containing five helical stems

HDV Requires low concentrations of divalent cations – play in an important role in folding or catalysis

Ribozymes: RNA Enzymes Regulation

Small Ribozymes (50 -150 nucleotides) Hammerhead, Hepatitis delta virus (HDV), hairpin Cleave only at specific location using base pairing and 3° interactions for alignment Catalysis does not require divalent metal ions

General Catalysis

General Catalysis Acid-Base

Hammerhead Ribozyme Blue= substrate Green-enzyme Red= cleavage site

Large Ribozymes (150+ nucleotides) �Group I and II introns, RNase. P �Catalysis requires divalent metal ions

Catalysis: Kinetics Ribozymes increase the rate of reaction by lowering the activation energy, just as DNA enzymes do

Catalysis: Kinetics Michaelis-Menton: applies to Ribozymes k 1 Rz + S Rz. S k 2 Rz + P k-1 Rz = ribozyme S = substrate P = product V = Vmax [S] / (Km + [S]) Michaelis-Menton Equation k-1+ k 2 Km= k 1 Michaelis Constant Effeciency = Kcat / Km

Catalysis: Kinetics

• Increase of ribozyme concentration does not change reaction rate • Temperature increase causes increase of reaction rate • p. H increase causes increase of reaction rate

References � http: //users. rcn. com/jkimball. ma. ultranet/Biology. Pages/R/Ribozymes. html � Scott, W. G. , Finch, J. T. and Klug, A. (1995). The Crystal Structure of an All-RNA Hammerhead Ribozyme: A Proposed Mechanism for Catalytic Cleavage. Cell. 81, 991 -1002. � Pley, H. W. , Flaherty, K. M. and Mc. Kay, D. B. (1994) Three-Dimensional Structure of a Hammerhead Ribozyme. Nature 372, 68 -74. � http: //www. sciencemag. org/content/282/5387/259. full � Alberts, Bruce, John H. Wilson, and Tim Hunt. Molecular Biology of the Cell. New York: Garland Science, 2008. Print. � Doherty, Elizabeth A. , and Jennifer A. Doudna. "Ribozyme Structures and Mechanisms. " Annual Review: Biophysics and Biomolecular Structure 30 (2001): 457 -75. Print. � Ferre´ -D’Amare, Adrian R. "Crystal Structure of a Hepatitis Delta. " Nature, 8 Oct. 1998. Web. <http: //cmgm. stanford. edu/biochem 201/Papers/doudna. pdf>. � Flugel, Rolf M. Chirality and Life a Short Introduction to the Early Phases of Chemical Evolution. Berlin: Springer, 2011. 23 -27. Print. � Reyes, Roberto. "Ribozymes: Molecular Scissors for Investigating Genetic Function. " Mad. Sci. Net: The 24 -hour Exploding Laboratory. University of Washington School of Medicine. Web. 16 Nov. 2011. <http: //www. madsci. org/dtm/ribozyme. html>. � Web. 19 Nov. 2011. <http: //www. pubmed. gov>.
- Slides: 37