Restriction Enzymes and Digestion What can you do










- Slides: 24

Restriction Enzymes and Digestion

What can you do with the Restriction Digestion and Analysis of Lambda DNA Kit? Understand the use of restriction enzymes as biotechnology tools and the mechanics of a restriction enzyme digest Become familiar with principals and techniques of agarose gel electrophoresis Estimate DNA fragment sizes from agarose gel data Understand the importance of restriction enzymes and their applications

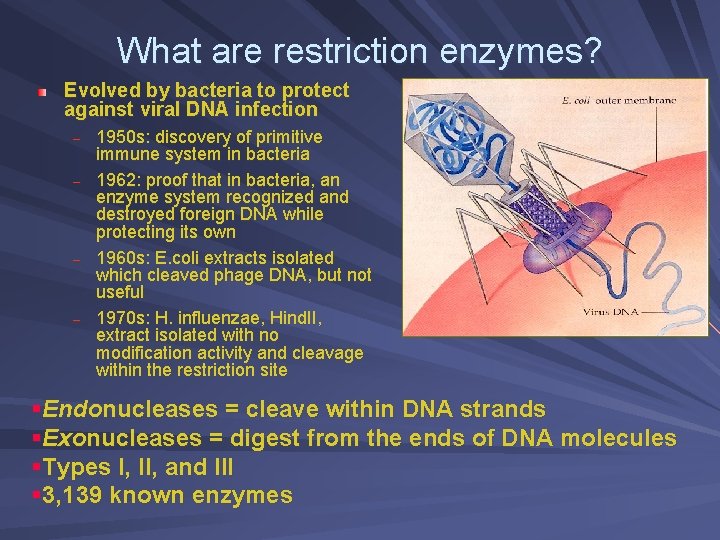
What are restriction enzymes? Evolved by bacteria to protect against viral DNA infection – – 1950 s: discovery of primitive immune system in bacteria 1962: proof that in bacteria, an enzyme system recognized and destroyed foreign DNA while protecting its own 1960 s: E. coli extracts isolated which cleaved phage DNA, but not useful 1970 s: H. influenzae, Hind. II, extract isolated with no modification activity and cleavage within the restriction site §Endonucleases = cleave within DNA strands §Exonucleases = digest from the ends of DNA molecules §Types I, II, and III § 3, 139 known enzymes

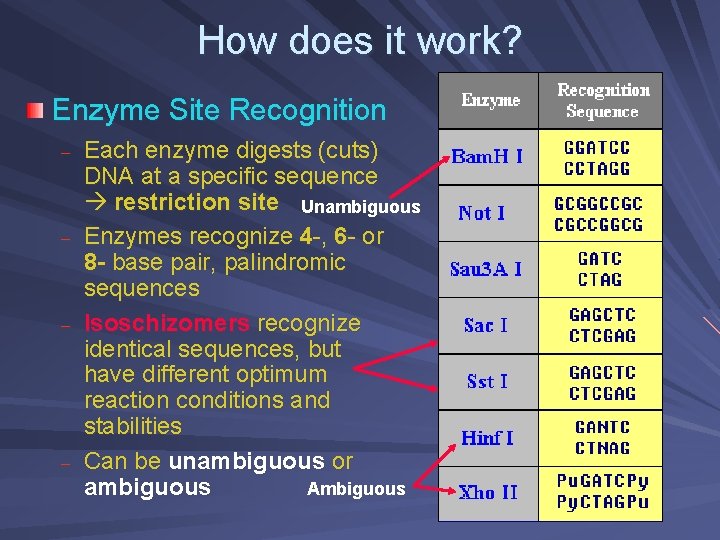
How does it work? Enzyme Site Recognition – – Each enzyme digests (cuts) DNA at a specific sequence restriction site Unambiguous Enzymes recognize 4 -, 6 - or 8 - base pair, palindromic sequences Isoschizomers recognize identical sequences, but have different optimum reaction conditions and stabilities Can be unambiguous or ambiguous Ambiguous

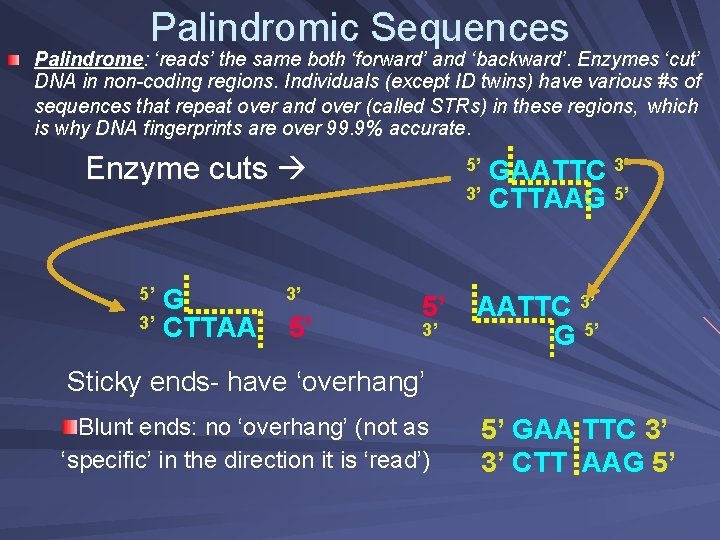
Palindromic Sequences Palindrome: ‘reads’ the same both ‘forward’ and ‘backward’. Enzymes ‘cut’ DNA in non-coding regions. Individuals (except ID twins) have various #s of sequences that repeat over and over (called STRs) in these regions, which is why DNA fingerprints are over 99. 9% accurate. Enzyme cuts G 3’ CTTAA 5’ 3’ 5’ GAATTC 3’ 3’ CTTAAG 5’ 5’ 5’ 3’ AATTC 3’ G 5’ Sticky ends- have ‘overhang’ Blunt ends: no ‘overhang’ (not as ‘specific’ in the direction it is ‘read’) 5’ GAA TTC 3’ 3’ CTT AAG 5’

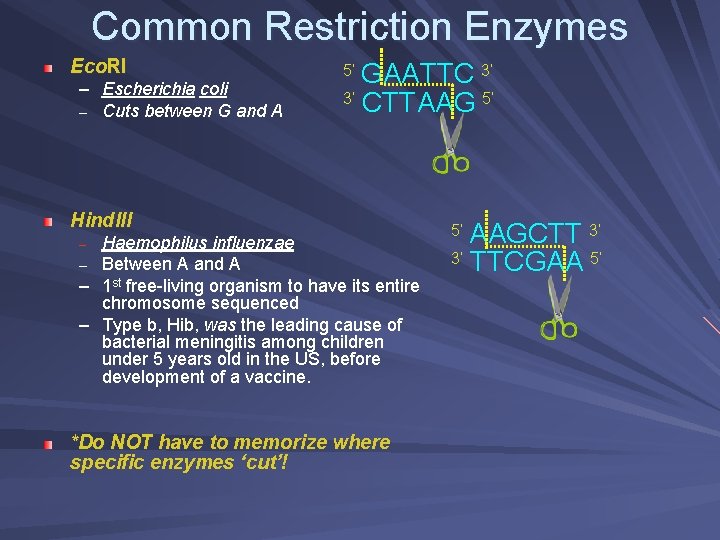
Common Restriction Enzymes Eco. RI – Escherichia coli – Cuts between G and A GAATTC 3’ 3’ CTTAAG 5’ 5’ Hind. III Haemophilus influenzae – Between A and A – 1 st free-living organism to have its entire chromosome sequenced – Type b, Hib, was the leading cause of bacterial meningitis among children under 5 years old in the US, before development of a vaccine. – *Do NOT have to memorize where specific enzymes ‘cut’! AAGCTT 3’ 3’ TTCGAA 5’ 5’

What is needed for restriction digestion? Template DNA, uncut DNA, often bacterial phage DNA Restriction enzyme(s), to cut template DNA Restriction Buffer, to provide optimal conditions for digestion Water Bath

Restriction Enzyme Digestion Restriction Buffer provides optimal conditions: Na. Cl provides correct ionic strength Tris-HCl provides the proper p. H Mg 2+ is an enzyme co-factor Different enzymes have different optimal buffers; Manufacturers package enzymes with buffers for ease of use

DNA Digestion Temperature Why incubate at 37 C? Body temperature is optimal for these and most other enzymes What happens if temperature is too hot or cool? Too hot = enzyme may be denatured, killed J Too cool= enzyme activity lowered, requiring longer digestion time G

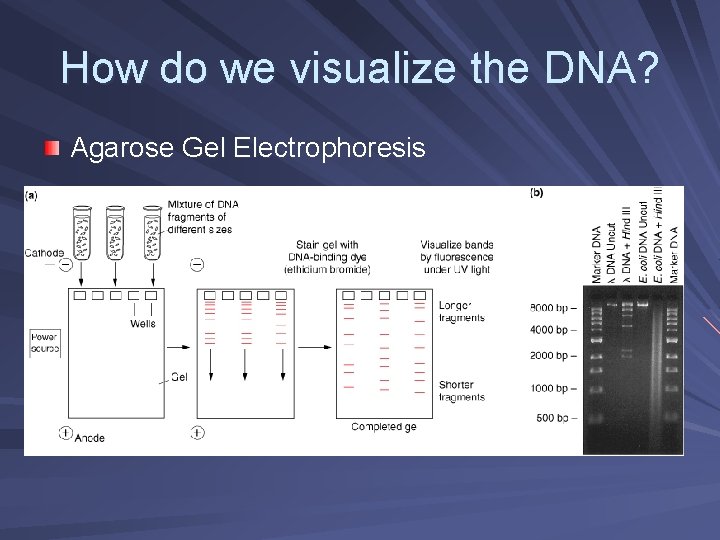
How do we visualize the DNA? Agarose Gel Electrophoresis

Agarose Gel Electrophoresis Electrolysis: the splitting of water using electricity – current splits water into hydrogen ions (H+) and hydroxyl ions (OH-) Electrophoresis: a method of separating charged molecules in an electrical field; DNA has an overall negative charge Used to separate DNA fragments by size

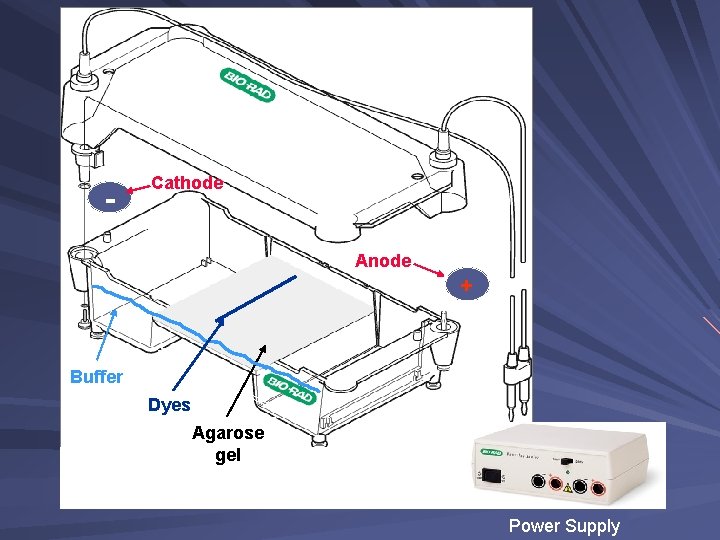
Components of an Electrophoresis System Power supply and chamber, a source of negatively charged particles with a cathode and anode Buffer, a fluid mixture of water and ions Agarose gel, a porous material that DNA migrates through Gel casting materials DNA ladder, mixture of DNA fragments of known lengths Loading dye, contains a dense material and allows visualization of DNA migration DNA Stain, allows visualizations of DNA fragments after electrophoresis

- Cathode Anode + Buffer Dyes Agarose gel Power Supply

Electrophoresis Buffer TAE (Tris-acetate-EDTA) and TBE (Trisborate-EDTA) are the most common buffers for duplex DNA Establish p. H and provide ions to support conductivity Concentration affects DNA migration – Use of water will produce no migraton – High buffer conc. could melt the agarose gel

Agarose Gel A porous material derived from red seaweed Acts as a sieve for separating DNA fragments; smaller fragments travel faster than large fragments 1% agarose – Plinko Model Concentration affects DNA migration – Low conc. = larger pores better resolution of larger DNA fragments – High conc. = smaller pores better resolution of smaller DNA fragments 2% agarose

Loading Dye DNA samples are loaded into a gel AFTER the tank has been filled with buffer, covering the gel Contains a dense substance, such as glycerol, to allow the sample to "fall" into the sample wells Contains one or two tracking dyes, which migrate in the gel and allow monitoring of how far the electrophoresis has proceeded.

DNA Staining Allows DNA visualization after gel electrophoresis Ethidium Bromide Bio-Safe DNA stains

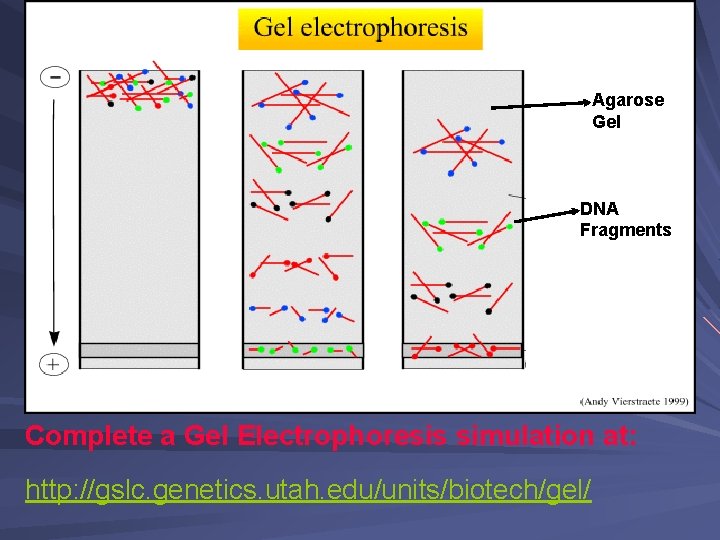
Agarose Gel DNA Fragments Complete a Gel Electrophoresis simulation at: http: //gslc. genetics. utah. edu/units/biotech/gel/

Restriction Enzyme Digest and Analysis Procedures

Actual Results of Restriction Enzyme Digestion Lane 1, DNA markers (Hind. III lambda digest) lane 2, uncut lambda DNA lane 3, lambda DNA digested with Pst. I lane 4, lambda DNA digested with Eco. RI lane 5, lambda DNA digested with Hind. III

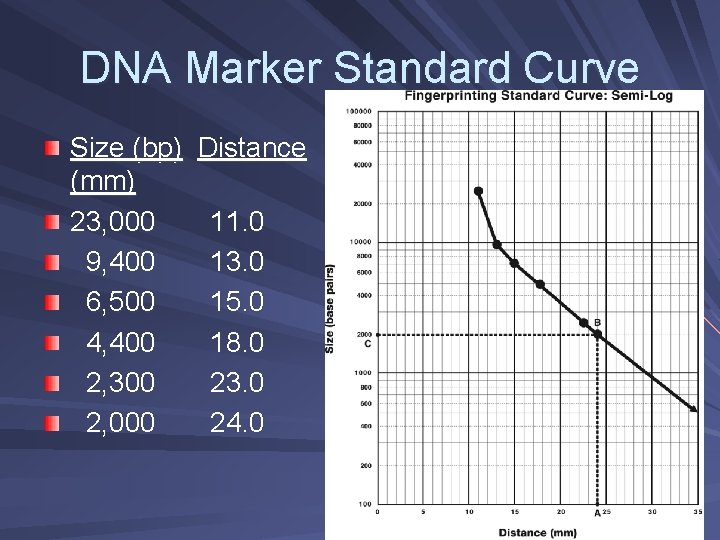
Analysis of DNA Fragments Determine restriction fragment sizes – Create standard curve using DNA marker – Measure distance traveled by restriction fragments – Determine size of DNA fragments

DNA Marker Standard Curve Size (bp) (mm) 23, 000 9, 400 6, 500 4, 400 2, 300 2, 000 Distance 11. 0 13. 0 15. 0 18. 0 23. 0 24. 0

Factors Affecting Restriction Enzyme Digestion Temperature, restriction enzymes are sensitive to prolonged periods of exposure to heat Cross contamination of restriction enzymes Buffer, optimum p. H Incubation temperature, maintain optimum temperature during restriction enzyme activity And Finally…Don’t forget to ADD your restriction enzyme to the reaction!!!

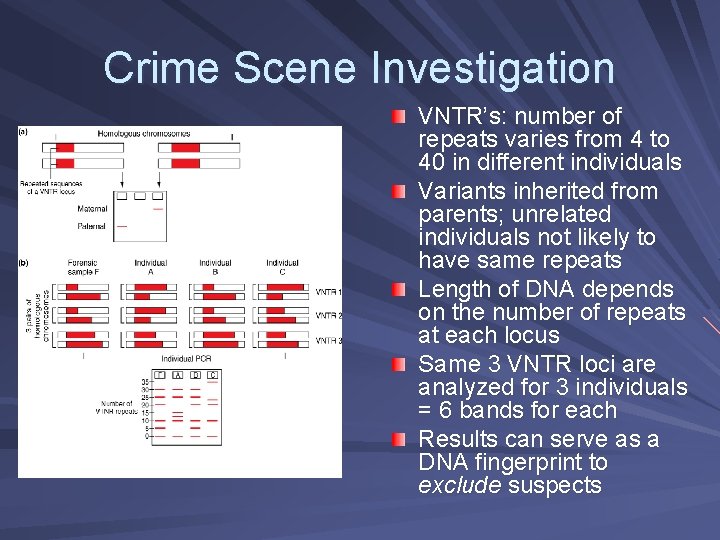
Crime Scene Investigation VNTR’s: number of repeats varies from 4 to 40 in different individuals Variants inherited from parents; unrelated individuals not likely to have same repeats Length of DNA depends on the number of repeats at each locus Same 3 VNTR loci are analyzed for 3 individuals = 6 bands for each Results can serve as a DNA fingerprint to exclude suspects