Resources at Hap Map Org Hap Map 3









- Slides: 53

Resources at Hap. Map. Org Hap. Map 3 Tutorial Marcela K. Tello-Ruiz Cold Spring Harbor Laboratory

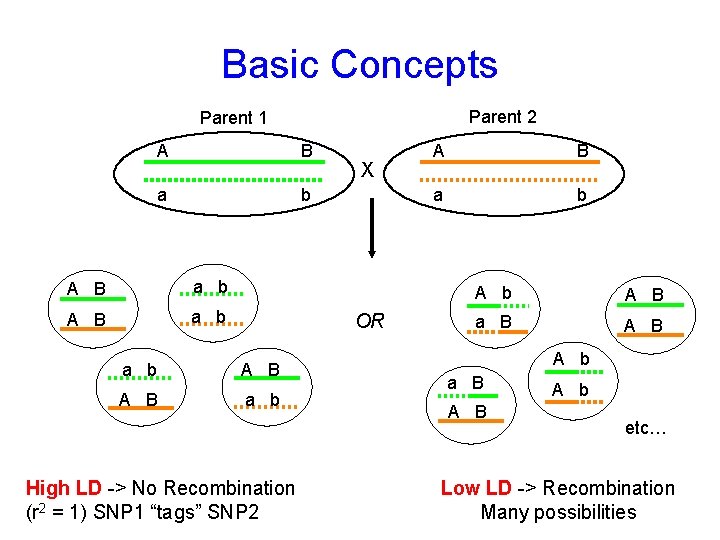
Basic Concepts Parent 2 Parent 1 A B a b X OR a b A B a b High LD -> No Recombination (r 2 = 1) SNP 1 “tags” SNP 2 A B a b A B a B A B A b etc… Low LD -> Recombination Many possibilities

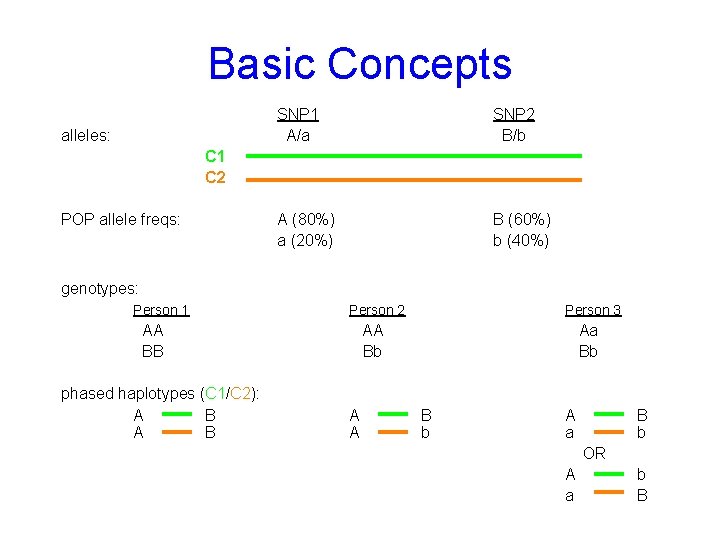
Basic Concepts alleles: SNP 1 A/a SNP 2 B/b A (80%) a (20%) B (60%) b (40%) C 1 C 2 POP allele freqs: genotypes: Person 1 Person 2 AA BB phased haplotypes (C 1/C 2): A B Person 3 AA Bb A A Aa Bb B b A a B b OR A a b B

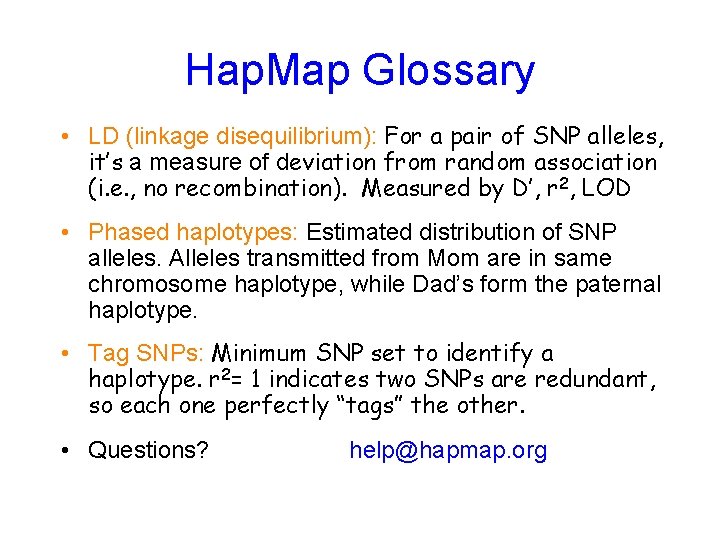
Hap. Map Glossary • LD (linkage disequilibrium): For a pair of SNP alleles, it’s a measure of deviation from random association (i. e. , no recombination). Measured by D’, r 2, LOD • Phased haplotypes: Estimated distribution of SNP alleles. Alleles transmitted from Mom are in same chromosome haplotype, while Dad’s form the paternal haplotype. • Tag SNPs: Minimum SNP set to identify a haplotype. r 2= 1 indicates two SNPs are redundant, so each one perfectly “tags” the other. • Questions? help@hapmap. org

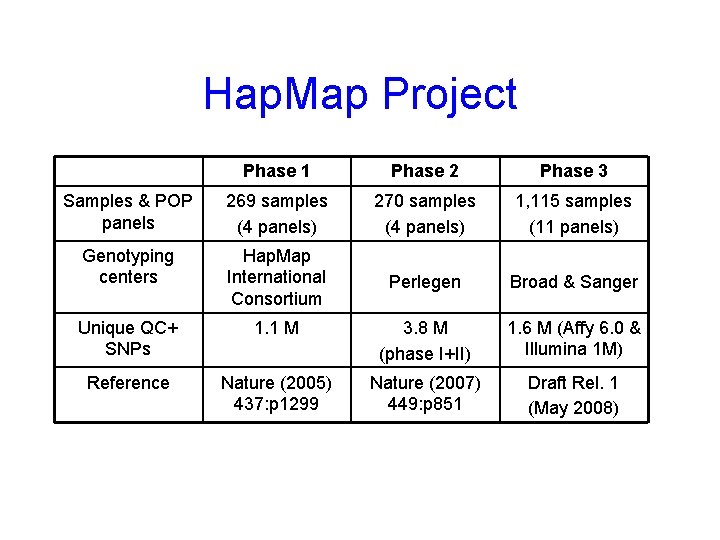
Hap. Map Project Phase 1 Phase 2 Phase 3 Samples & POP panels 269 samples (4 panels) 270 samples (4 panels) 1, 115 samples (11 panels) Genotyping centers Hap. Map International Consortium Perlegen Broad & Sanger Unique QC+ SNPs 1. 1 M 3. 8 M (phase I+II) 1. 6 M (Affy 6. 0 & Illumina 1 M) Reference Nature (2005) 437: p 1299 Nature (2007) 449: p 851 Draft Rel. 1 (May 2008)

Release Notes • Phase 1+2: Latest Release #24, October 2008 (NCBI build 36): 3. 9 M unique QC+ SNPs -- > 1 SNP/700 bp http: //ftp. hapmap. org/00 README. releasenotes_rel 24 – Added back chr. X SNPs dropped in previous releases – Corrected allele flips from rel#23 a • Phase 3: Draft release #1 (NCBI build 36) http: //ftp. hapmap. org/genotypes/2008 -07_phase. III/00 README. txt – Hap. Map 3 sites @ Broad Institute, Sanger Center and Baylor College

Phase 3 Samples * Population is made of family trios

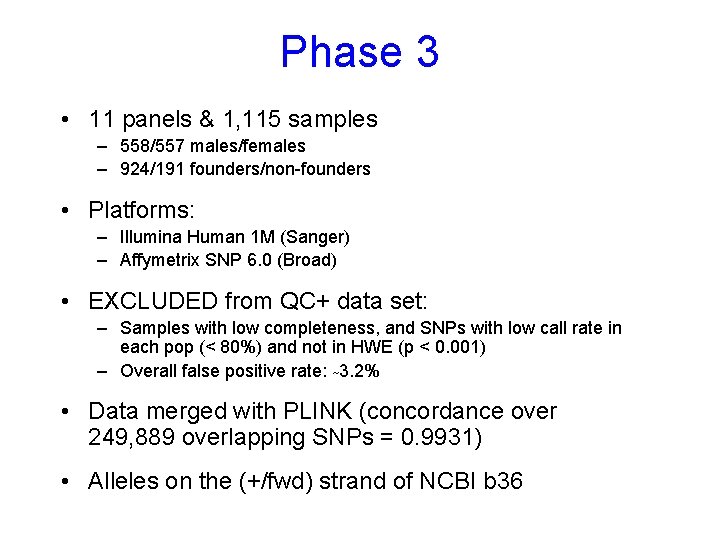
Phase 3 • 11 panels & 1, 115 samples – 558/557 males/females – 924/191 founders/non-founders • Platforms: – Illumina Human 1 M (Sanger) – Affymetrix SNP 6. 0 (Broad) • EXCLUDED from QC+ data set: – Samples with low completeness, and SNPs with low call rate in each pop (< 80%) and not in HWE (p < 0. 001) – Overall false positive rate: ~3. 2% • Data merged with PLINK (concordance over 249, 889 overlapping SNPs = 0. 9931) • Alleles on the (+/fwd) strand of NCBI b 36

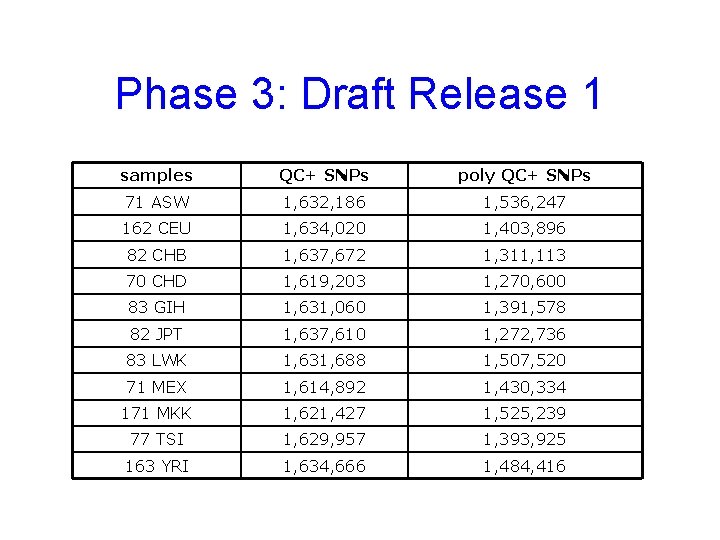
Phase 3: Draft Release 1 samples QC+ SNPs poly QC+ SNPs 71 ASW 1, 632, 186 1, 536, 247 162 CEU 1, 634, 020 1, 403, 896 82 CHB 1, 637, 672 1, 311, 113 70 CHD 1, 619, 203 1, 270, 600 83 GIH 1, 631, 060 1, 391, 578 82 JPT 1, 637, 610 1, 272, 736 83 LWK 1, 631, 688 1, 507, 520 71 MEX 1, 614, 892 1, 430, 334 171 MKK 1, 621, 427 1, 525, 239 77 TSI 1, 629, 957 1, 393, 925 163 YRI 1, 634, 666 1, 484, 416

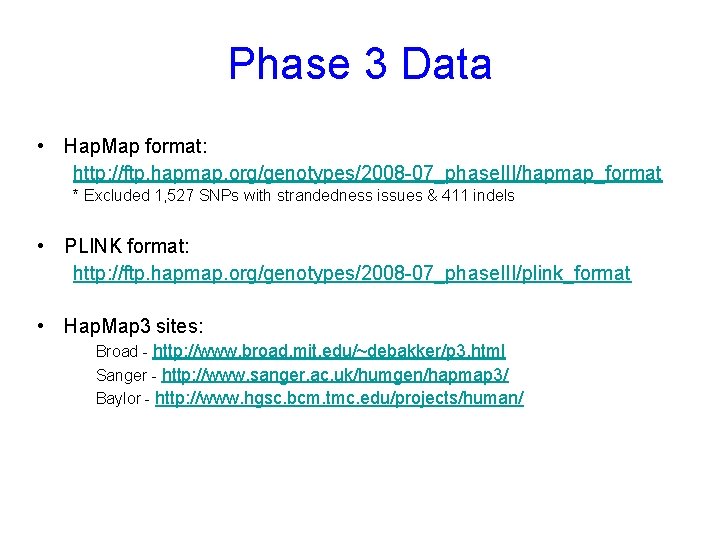
Phase 3 Data • Hap. Map format: http: //ftp. hapmap. org/genotypes/2008 -07_phase. III/hapmap_format * Excluded 1, 527 SNPs with strandedness issues & 411 indels • PLINK format: http: //ftp. hapmap. org/genotypes/2008 -07_phase. III/plink_format • Hap. Map 3 sites: Broad - http: //www. broad. mit. edu/~debakker/p 3. html Sanger - http: //www. sanger. ac. uk/humgen/hapmap 3/ Baylor - http: //www. hgsc. bcm. tmc. edu/projects/human/

Goals of This Tutorial This tutorial will show you how to: • Find Hap. Map 3 SNPs near a gene or region of interest (ROI) – – – • Visualize allele frequencies in Hap. Map 3 populations Download SNP genotypes in ROI for use in Haploview 4. 1 Identify GWA hits in the vicinity of ROI & visualize in the context of all chromosomes (karyogram) Add custom data onto the GWAs karyogram Add custom tracks of association data onto ROI Create publication-quality images Download the entire Hap. Map 3 data set in bulk – Distinguish genotype data in PLINK and Hap. Map formats • Visualize LD patterns, find tag SNPs, impute genotypes using release #24 (phase 1+2) • Generate customized extracts of the entire dataset using Hap. Mart

1: Surf to the Hap. Map Browser 1 a. Go to www. hapmap. org 1 b. Select “Hap. Map phase 3”

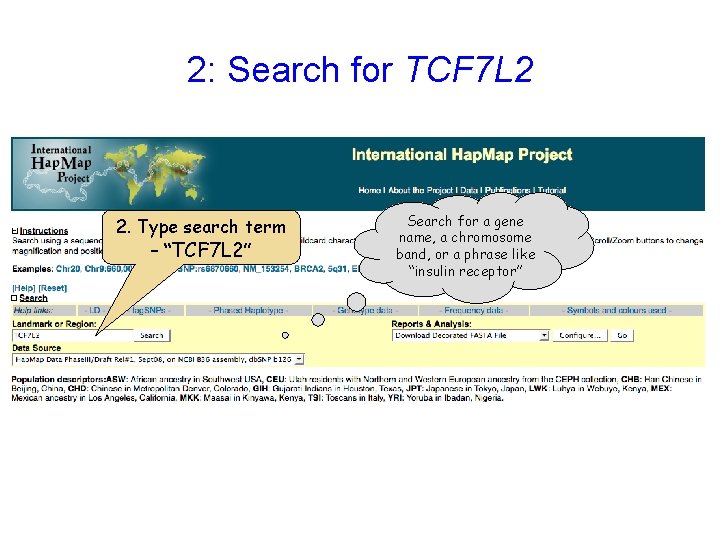
2: Search for TCF 7 L 2 2. Type search term – “TCF 7 L 2” Search for a gene name, a chromosome band, or a phrase like “insulin receptor”

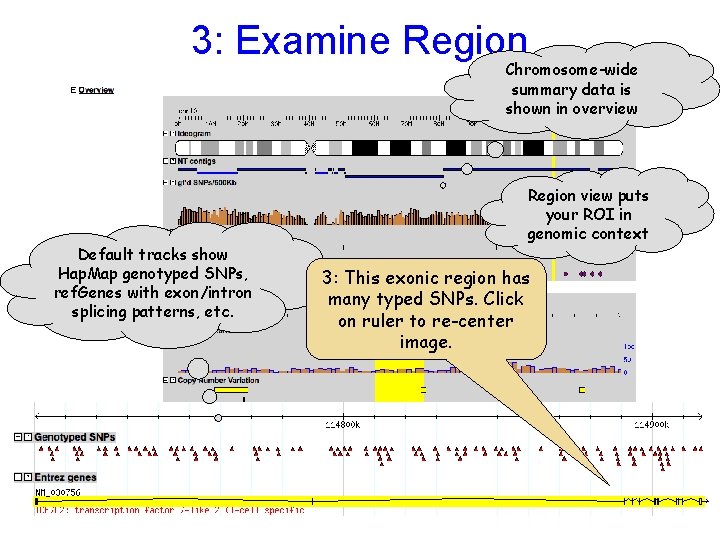
3: Examine Region Chromosome-wide summary data is shown in overview Default tracks show Hap. Map genotyped SNPs, ref. Genes with exon/intron splicing patterns, etc. Region view puts your ROI in genomic context 3: This exonic region has many typed SNPs. Click on ruler to re-center image.

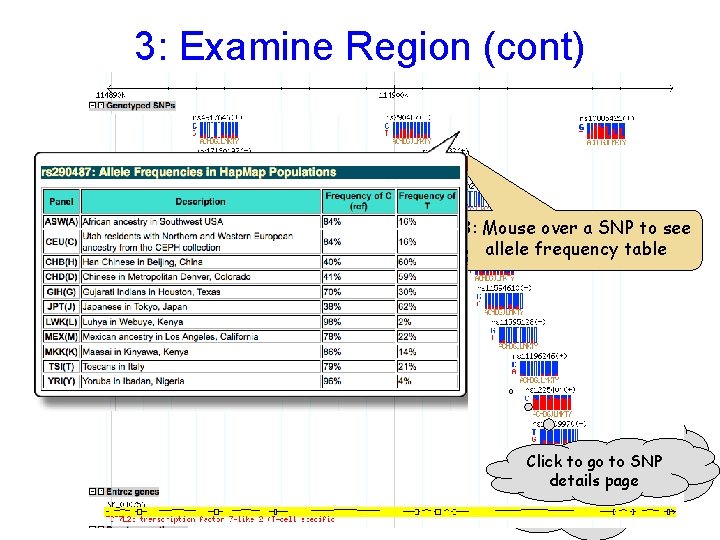
3: Examine Region (cont) Use the Scroll/Zoom buttons and menu to change position & magnification 3: Mouse over a SNP to see allele frequency table As you zoom in Click to gothe to SNP further, details pageto display changes include more detail

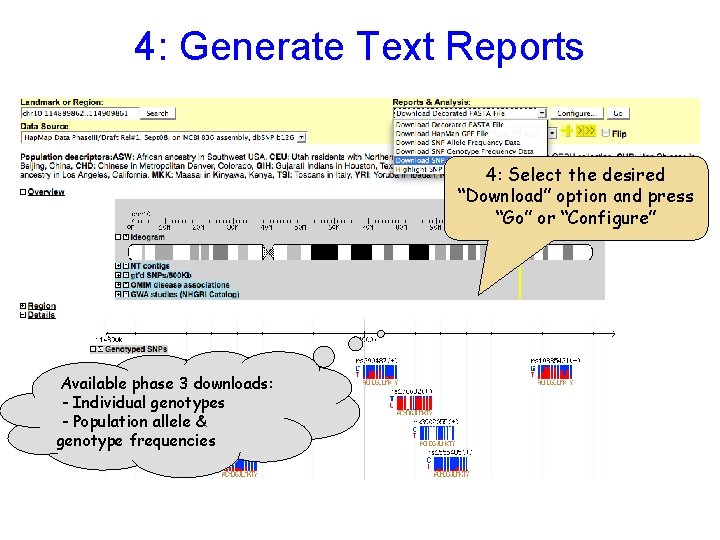
4: Generate Text Reports 4: Select the desired “Download” option and press “Go” or “Configure” Available phase 3 downloads: - Individual genotypes - Population allele & genotype frequencies

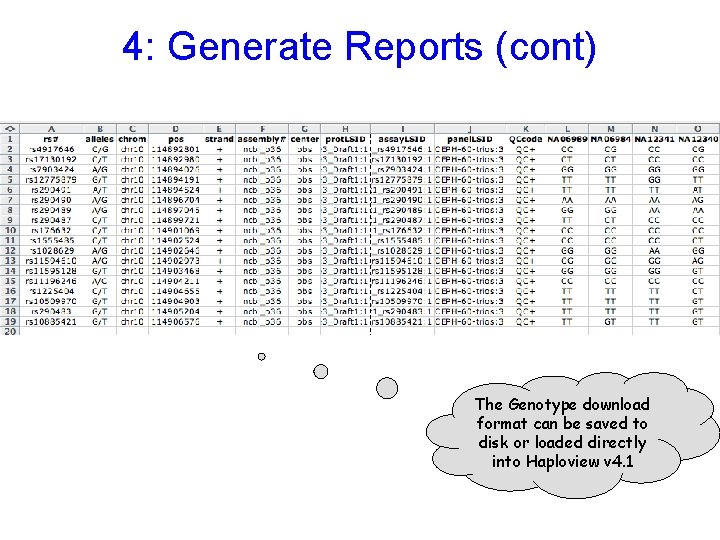
4: Generate Reports (cont) The Genotype download format can be saved to disk or loaded directly into Haploview v 4. 1

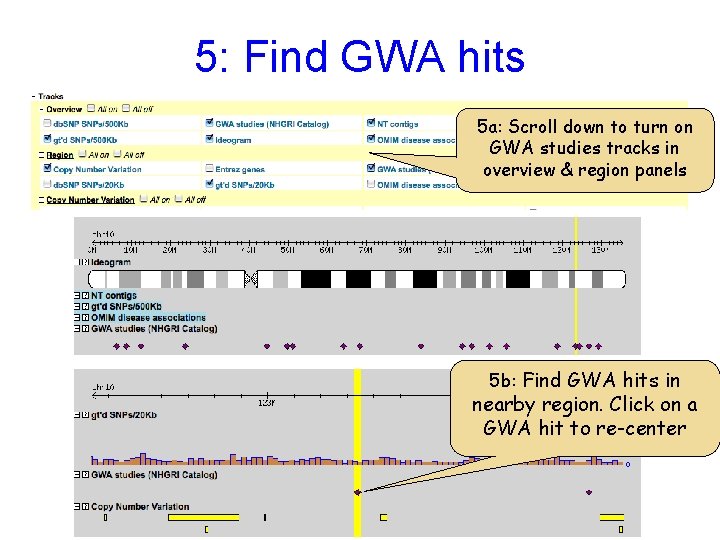
5: Find GWA hits 5 a: Scroll down to turn on GWA studies tracks in overview & region panels 5 b: Find GWA hits in nearby region. Click on a GWA hit to re-center

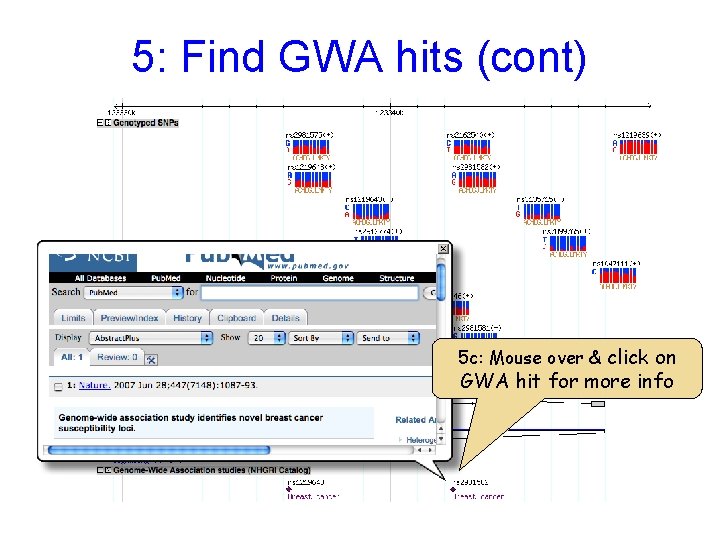
5: Find GWA hits (cont) 5 c: Mouse over & click on GWA hit for more info

6: Examine GWA hits in entire genome 6: From www. hapmap. org, select “Karyogram”

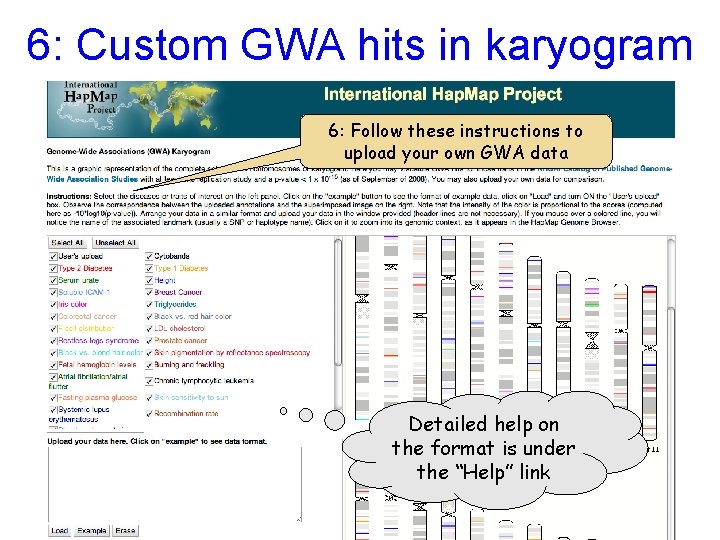
6: Custom GWA hits in karyogram 6: Follow these instructions to upload your own GWA data Detailed help on the format is under the “Help” link

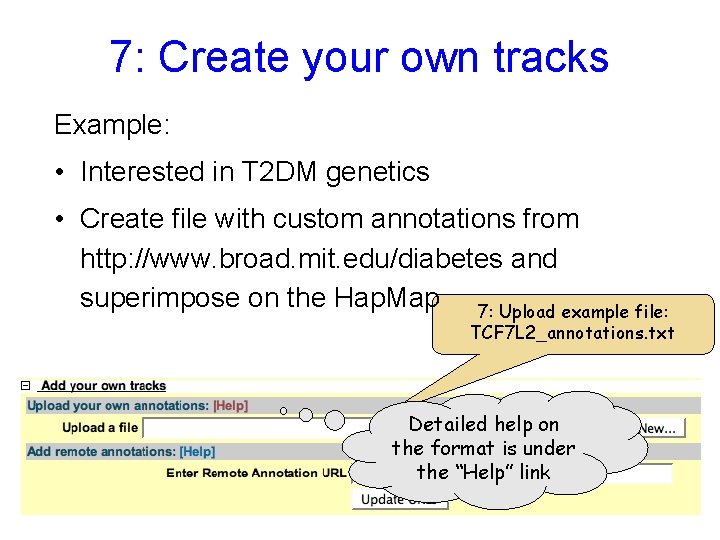
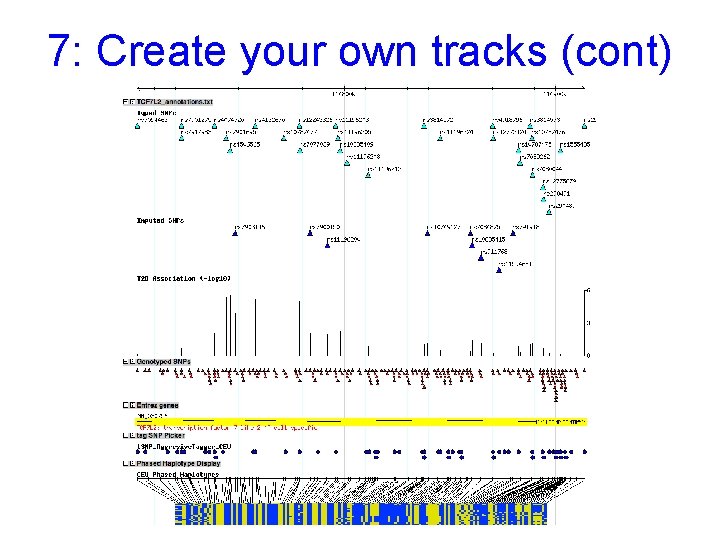
7: Create your own tracks Example: • Interested in T 2 DM genetics • Create file with custom annotations from http: //www. broad. mit. edu/diabetes and superimpose on the Hap. Map 7: Upload example file: TCF 7 L 2_annotations. txt Detailed help on the format is under the “Help” link

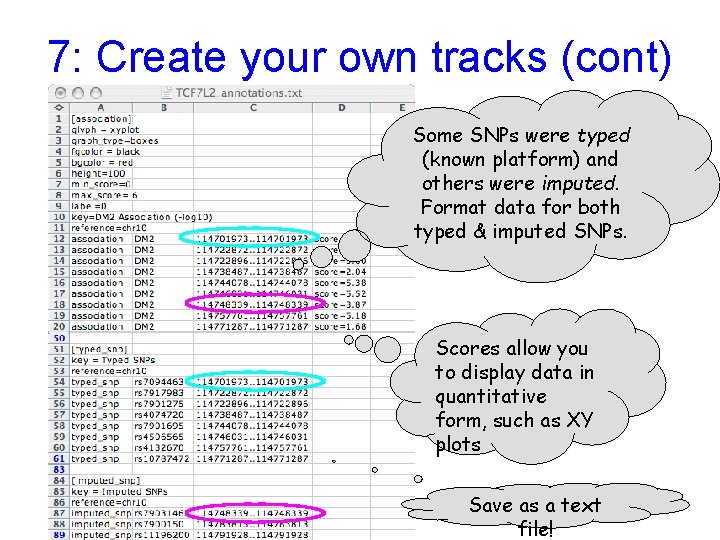
7: Create your own tracks (cont) Some SNPs were typed (known platform) and others were imputed. Format data for both typed & imputed SNPs. Scores allow you to display data in quantitative form, such as XY plots Save as a text file!

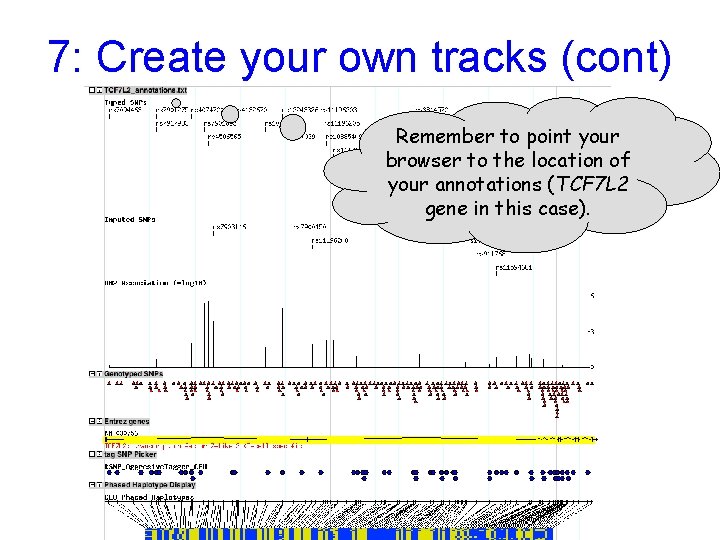
7: Create your own tracks (cont) Remember to point your browser to the location of your annotations (TCF 7 L 2 gene in this case).

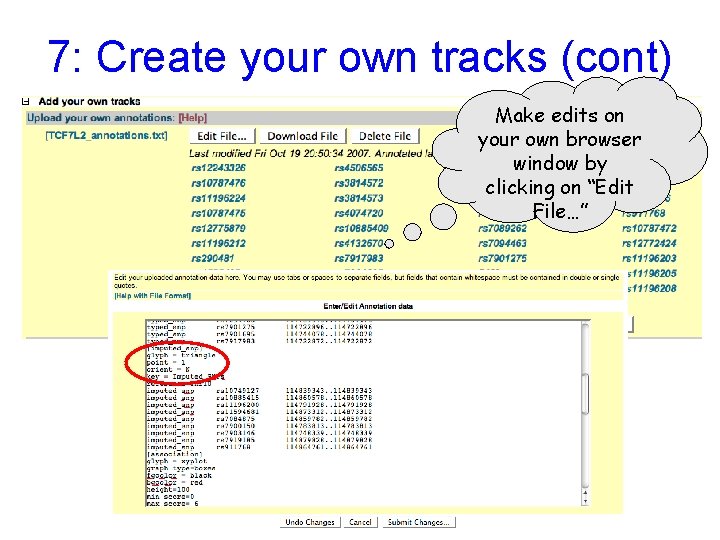
7: Create your own tracks (cont) Make edits on your own browser window by clicking on “Edit File…”

7: Create your own tracks (cont)

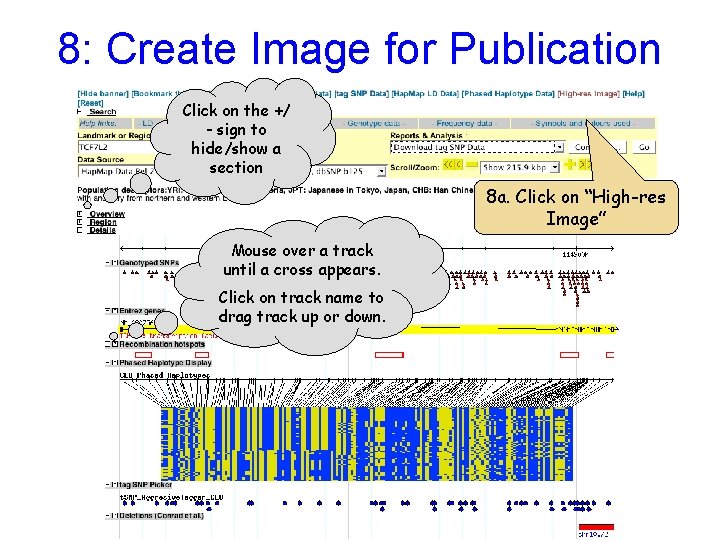
8: Create Image for Publication Click on the +/ - sign to hide/show a section 8 a. Click on “High-res Image” Mouse over a track until a cross appears. Click on track name to drag track up or down.

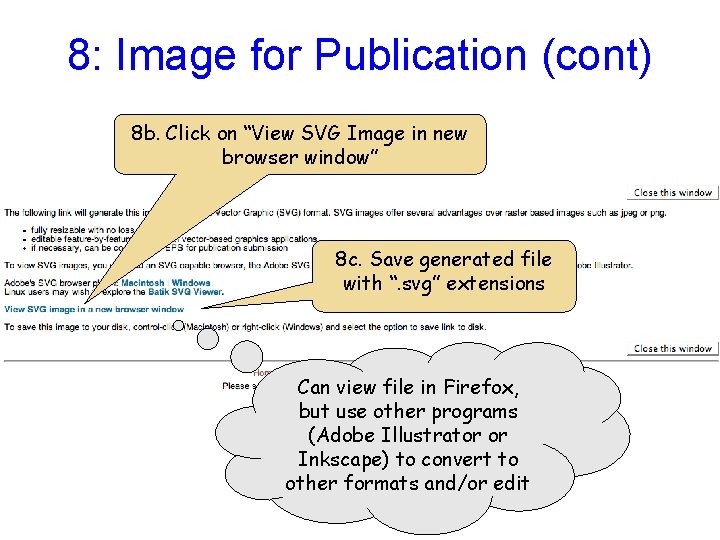
8: Image for Publication (cont) 8 b. Click on “View SVG Image in new browser window” 8 c. Save generated file with “. svg” extensions Can view file in Firefox, but use other programs (Adobe Illustrator or Inkscape) to convert to other formats and/or edit

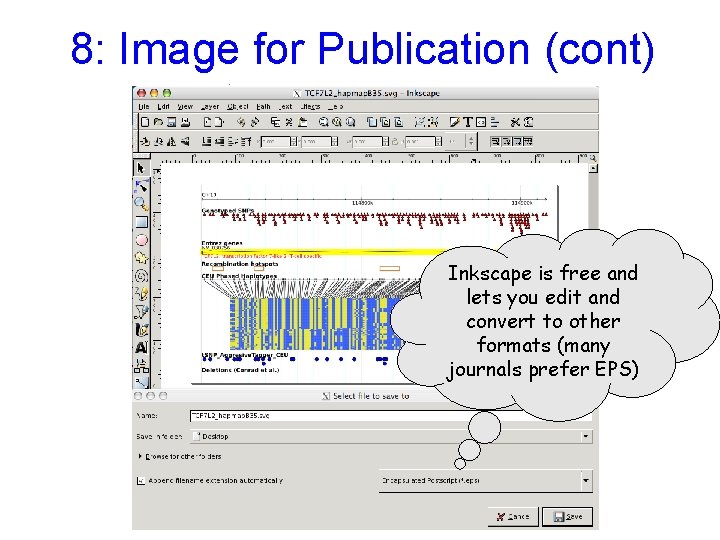
8: Image for Publication (cont) Inkscape is free and lets you edit and convert to other formats (many journals prefer EPS)

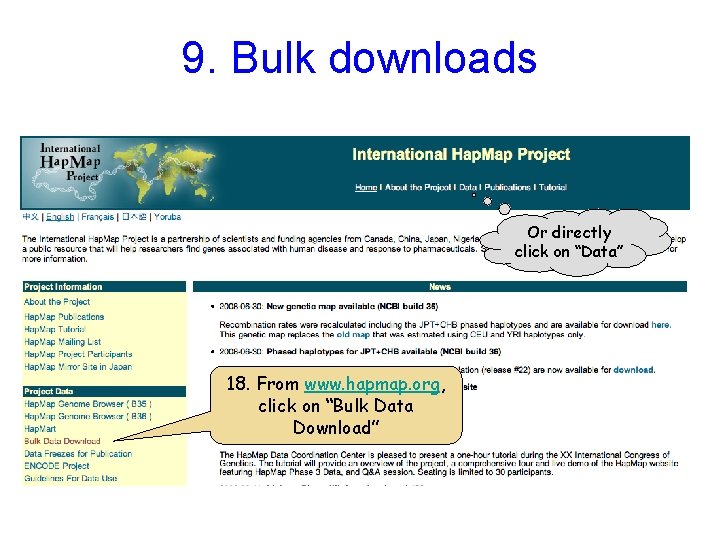
9. Bulk downloads Or directly click on “Data” 18. From www. hapmap. org, click on “Bulk Data Download”

9. Bulk downloads Download the entire Hap. Map 3 data set to your own computer Hap. Map 3 genotypes & frequencies 9 a. Select “Genotypes” Analytic results (LD & phased haplotype data available for Hap. Map 3) Your own copy of the Hap. Map Browser Protocols & assay design Hap. Map Samples Also available at http: //ftp. hapmap. org

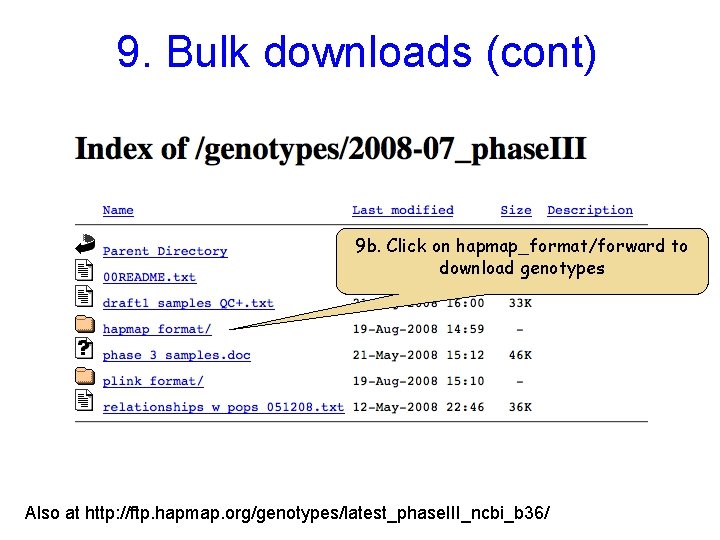
9. Bulk downloads (cont) 9 b. Click on hapmap_format/forward to download genotypes Also at http: //ftp. hapmap. org/genotypes/latest_phase. III_ncbi_b 36/

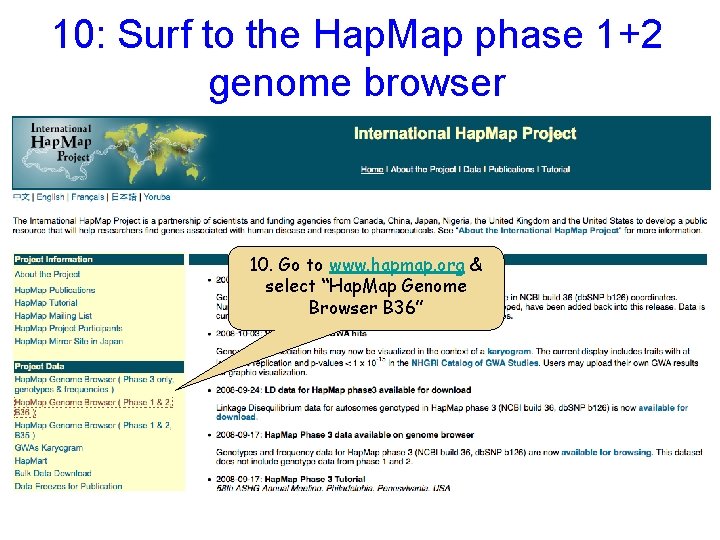
10: Surf to the Hap. Map phase 1+2 genome browser 10. Go to www. hapmap. org & select “Hap. Map Genome Browser B 36”

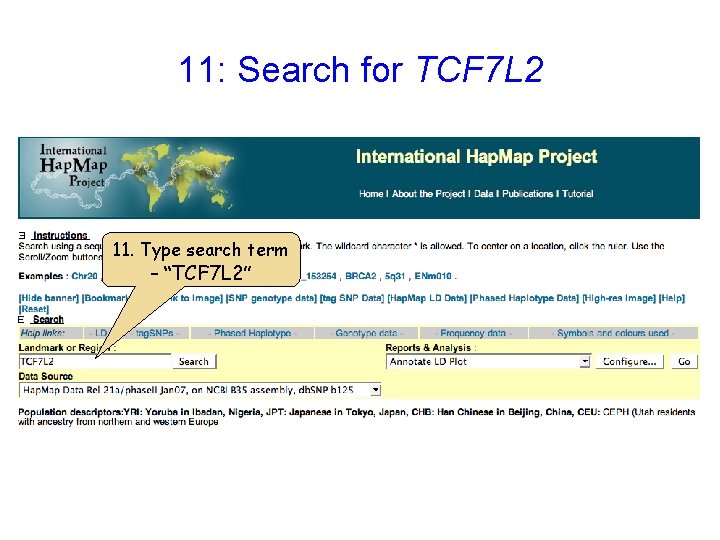
11: Search for TCF 7 L 2 11. Type search term – “TCF 7 L 2”

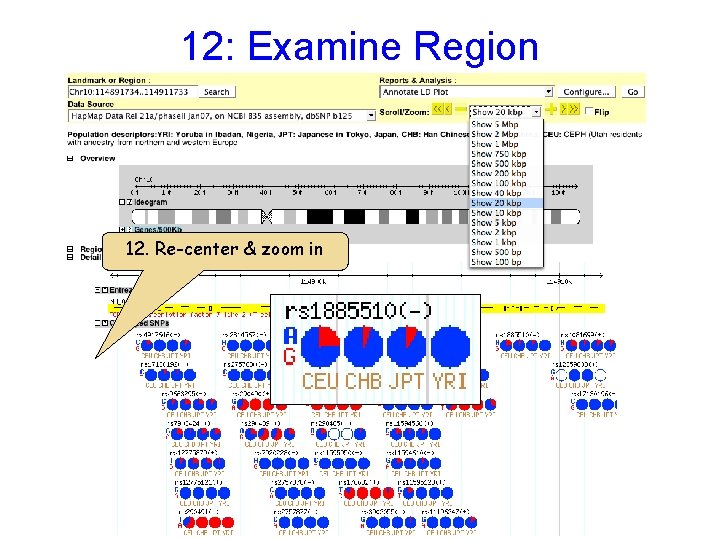
12: Examine Region 12. Re-center & zoom in

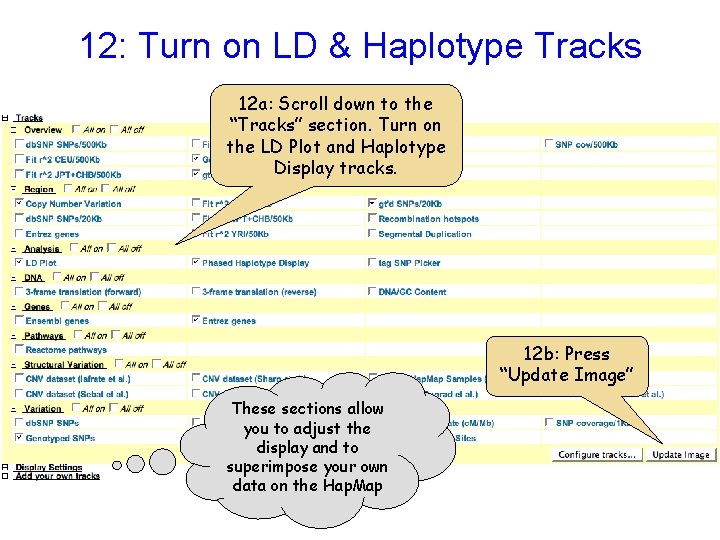
12: Turn on LD & Haplotype Tracks 12 a: Scroll down to the “Tracks” section. Turn on the LD Plot and Haplotype Display tracks. 12 b: Press “Update Image” These sections allow you to adjust the display and to superimpose your own data on the Hap. Map

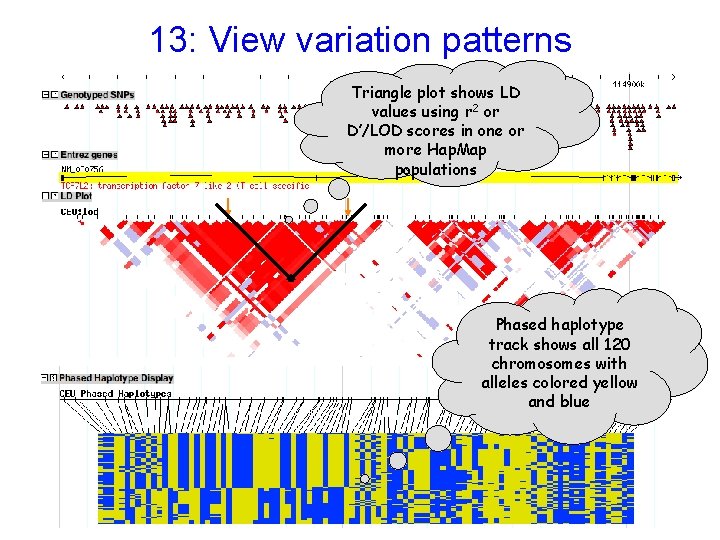
13: View variation patterns Triangle plot shows LD values using r 2 or D’/LOD scores in one or more Hap. Map populations Phased haplotype track shows all 120 chromosomes with alleles colored yellow and blue

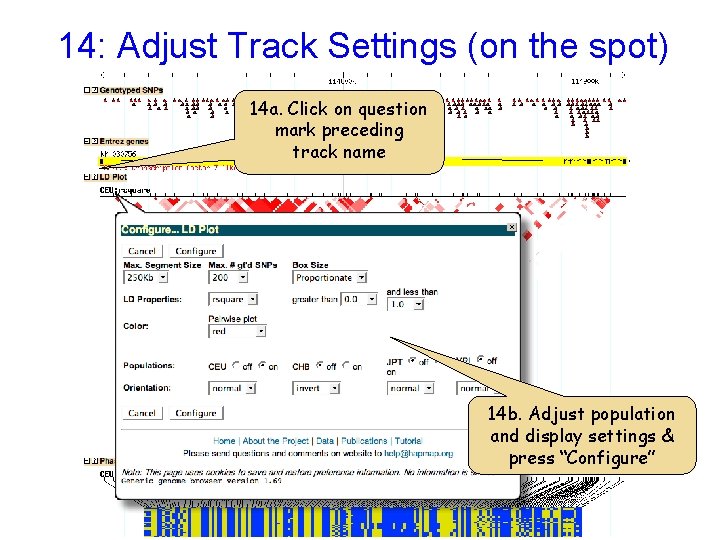
14: Adjust Track Settings (on the spot) 14 a. Click on question mark preceding track name 14 b. Adjust population and display settings & press “Configure”

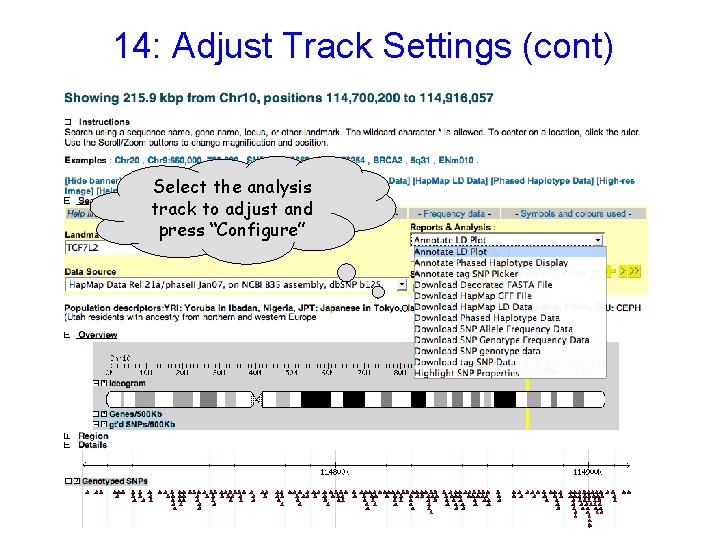
14: Adjust Track Settings (cont) Select the analysis track to adjust and press “Configure”

15: Turn on Tag SNP Track 15: Activate the “tag SNP Picker” and press “Update Image”

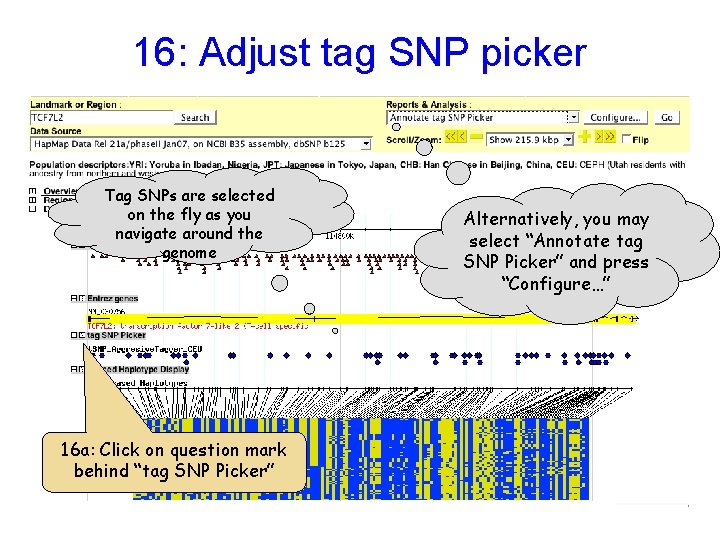
16: Adjust tag SNP picker Tag SNPs are selected on the fly as you navigate around the genome 16 a: Click on question mark behind “tag SNP Picker” Alternatively, you may select “Annotate tag SNP Picker” and press “Configure…”

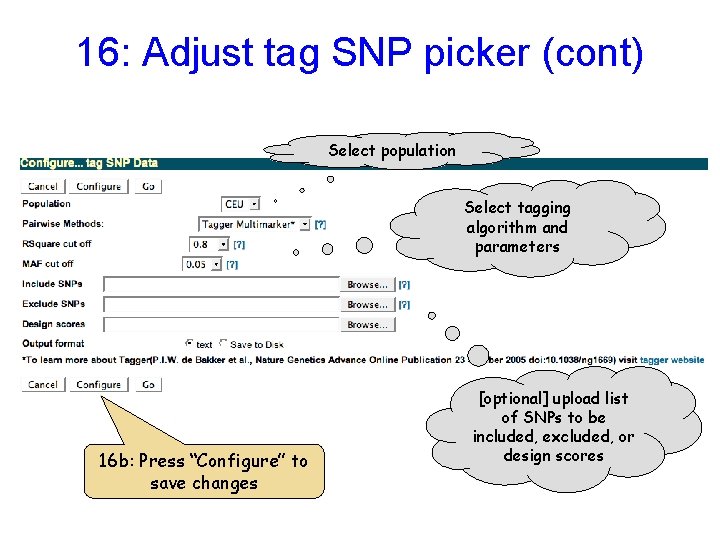
16: Adjust tag SNP picker (cont) Select population Select tagging algorithm and parameters 16 b: Press “Configure” to save changes [optional] upload list of SNPs to be included, excluded, or design scores

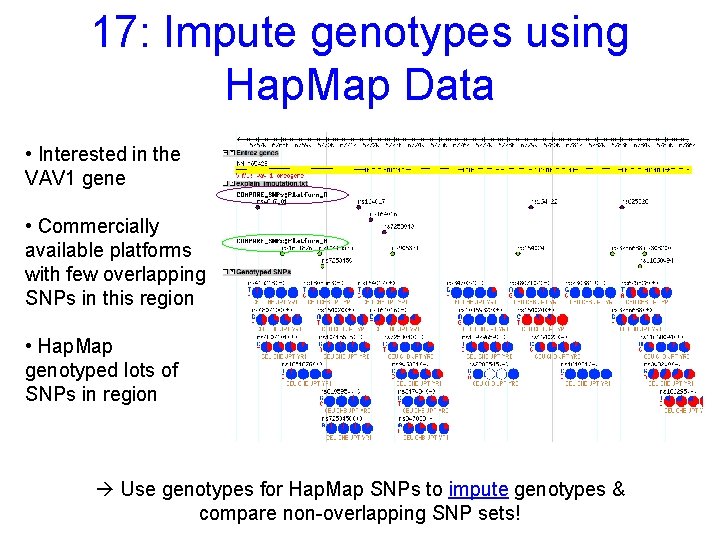
17: Impute genotypes using Hap. Map Data • Interested in the VAV 1 gene • Commercially available platforms with few overlapping SNPs in this region • Hap. Map genotyped lots of SNPs in region à Use genotypes for Hap. Map SNPs to impute genotypes & compare non-overlapping SNP sets!

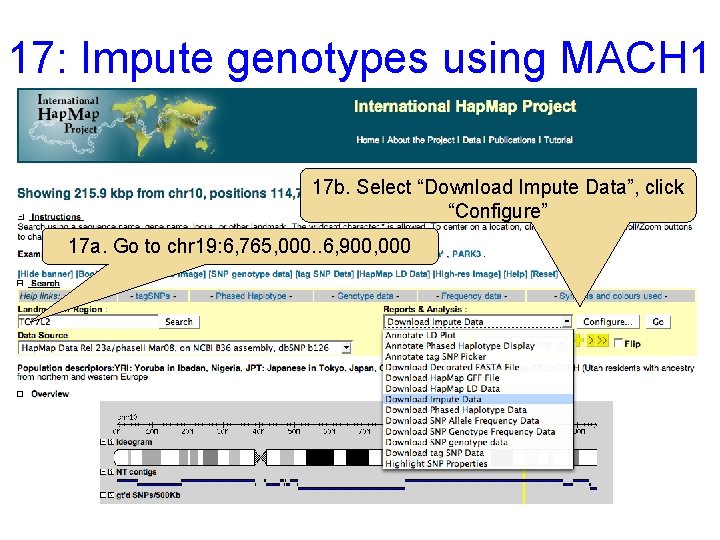
17: Impute genotypes using MACH 1 17 b. Select “Download Impute Data”, click “Configure” 17 a. Go to chr 19: 6, 765, 000. . 6, 900, 000

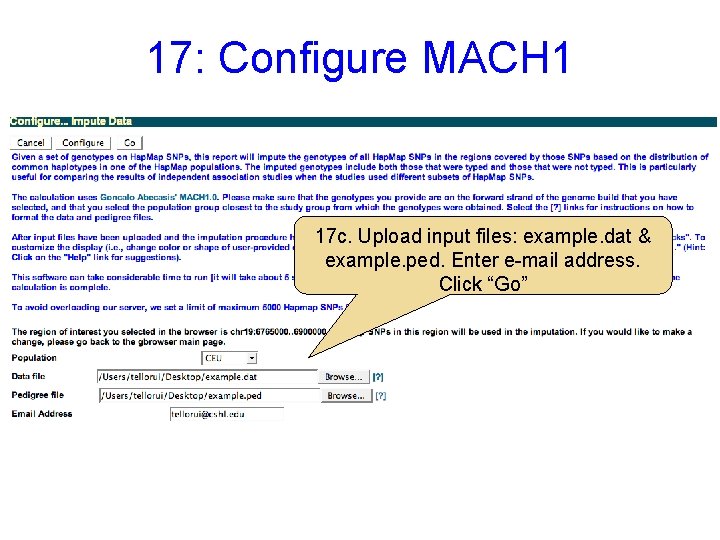
17: Configure MACH 1 17 c. Upload input files: example. dat & example. ped. Enter e-mail address. Click “Go”

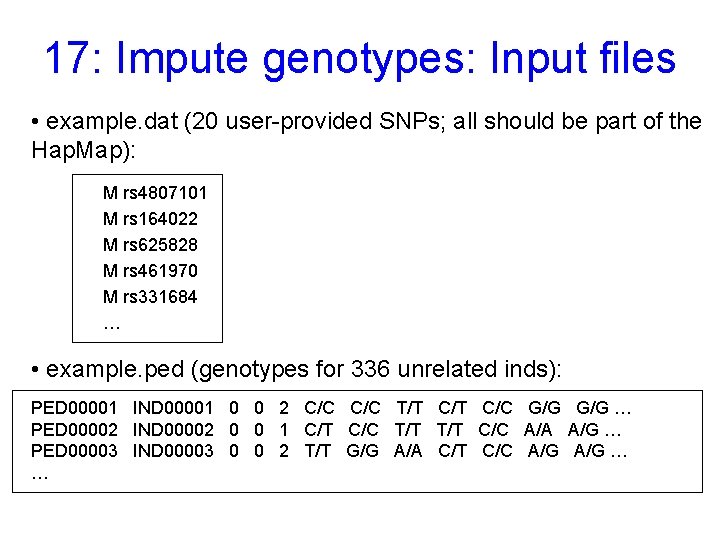
17: Impute genotypes: Input files • example. dat (20 user-provided SNPs; all should be part of the Hap. Map): M rs 4807101 M rs 164022 M rs 625828 M rs 461970 M rs 331684 … • example. ped (genotypes for 336 unrelated inds): PED 00001 IND 00001 0 0 2 C/C T/T C/C G/G … PED 00002 IND 00002 0 0 1 C/T C/C T/T C/C A/A A/G … PED 00003 IND 00003 0 0 2 T/T G/G A/A C/T C/C A/G … …

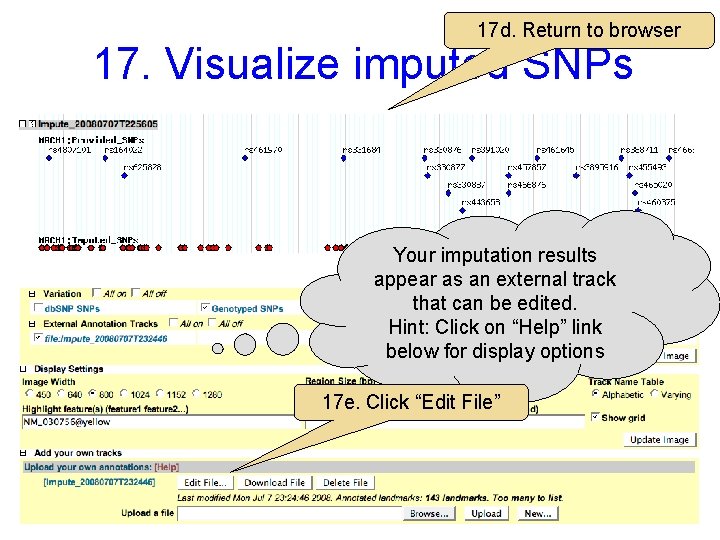
17 d. Return to browser 17. Visualize imputed SNPs Your imputation results appear as an external track that can be edited. Hint: Click on “Help” link below for display options 17 e. Click “Edit File”

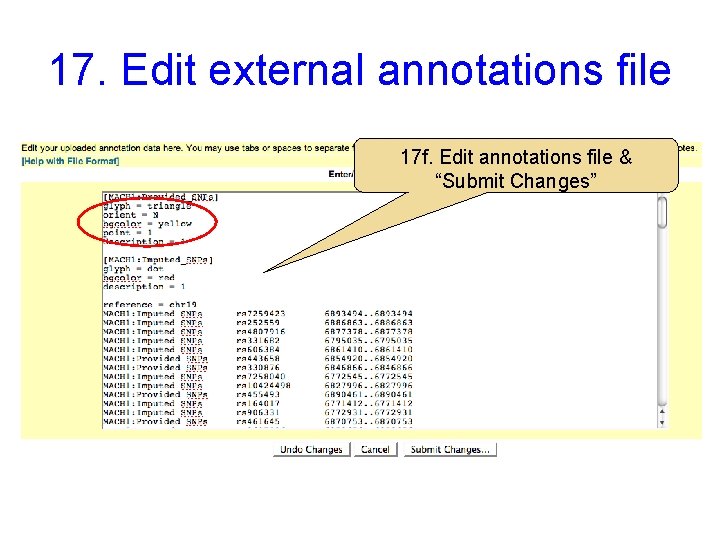
17. Edit external annotations file 17 f. Edit annotations file & “Submit Changes”

17. Edit external annotations file

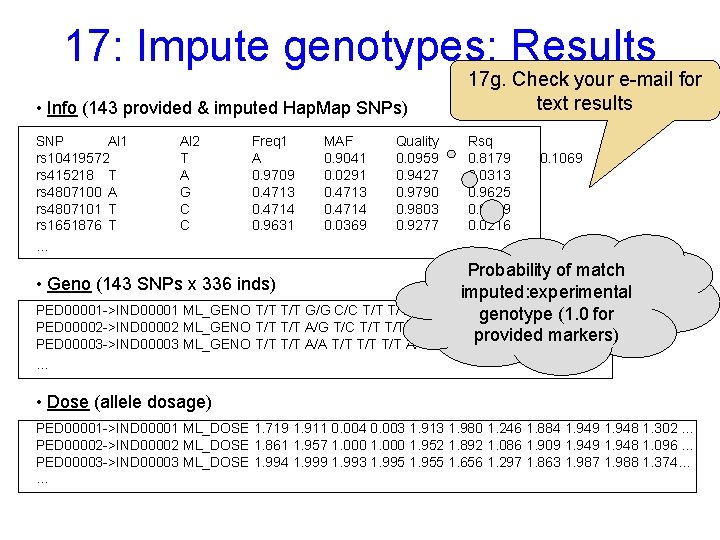
17: Impute genotypes: Results • Info (143 provided & imputed Hap. Map SNPs) SNP Al 1 rs 10419572 rs 415218 T rs 4807100 A rs 4807101 T rs 1651876 T … Al 2 T A G C C Freq 1 A 0. 9709 0. 4713 0. 4714 0. 9631 MAF 0. 9041 0. 0291 0. 4713 0. 4714 0. 0369 Quality 0. 0959 0. 9427 0. 9790 0. 9803 0. 9277 17 g. Check your e-mail for text results Rsq 0. 8179 0. 0313 0. 9625 0. 9649 0. 0216 0. 1069 Probability of match imputed: experimental PED 00001 ->IND 00001 ML_GENO T/T G/G C/C T/T A/T G/G A/A T/T T/C …(1. 0 for genotype PED 00002 ->IND 00002 ML_GENO T/T A/G T/C T/T A/T G/G A/A T/T T/C … provided markers) PED 00003 ->IND 00003 ML_GENO T/T A/A T/T T/T A/T G/G A/A T/T … • Geno (143 SNPs x 336 inds) … • Dose (allele dosage) PED 00001 ->IND 00001 ML_DOSE 1. 719 1. 911 0. 004 0. 003 1. 913 1. 980 1. 246 1. 884 1. 949 1. 948 1. 302 … PED 00002 ->IND 00002 ML_DOSE 1. 861 1. 957 1. 000 1. 952 1. 892 1. 086 1. 909 1. 948 1. 096 … PED 00003 ->IND 00003 ML_DOSE 1. 994 1. 999 1. 993 1. 995 1. 955 1. 656 1. 297 1. 863 1. 987 1. 988 1. 374… …

18. Use Hap. Mart to Generate Extracts of the Hap. Map Dataset Find all Hap. Map characterized SNPs that: 1. Have a MAF > 0. 20 in the Yoruban population panel (YRI) 2. Cause a nonsynonymous amino acid change 3. Were typed by Perlegen

Further Information • Hap. Map Publications & Guidelines http: //hapmap. cshl. org/publications. html. en • Past tutorials & user’s guide to Hap. Map. org http: //www. hapmap. org/tutorials. html. en • Questions? help@hapmap. org

Hap. Map DCC Present Members (CSHL) Lincoln Stein Marcela K. Tello-Ruiz Zhenyuan Lu Wei Zhao Hap. Map DCC Former Members Lalitha Krishnan Albert Vernon Smith Gudmundur Thorisson Fiona Cunningham