Research Techniques Made Simple Flow Cytometry II Mass

- Slides: 9

Research Techniques Made Simple Flow Cytometry II: Mass and Imaging Cytometry Hung Doan 1, 2, Garrett M. Chinn 3, Richard R. Jahan-Tigh 1, 2 Department of Dermatology, University of Texas, Houston Medical School, Houston, TX, USA 2 Division of Dermatology, Department of Medicine, University of Texas MD Anderson Cancer Center, Houston, TX, USA 3 Division of General Internal Medicine, Department of Medicine, Harvard Medical School, Massachusetts General Hospital, Boston, Massachusetts 1

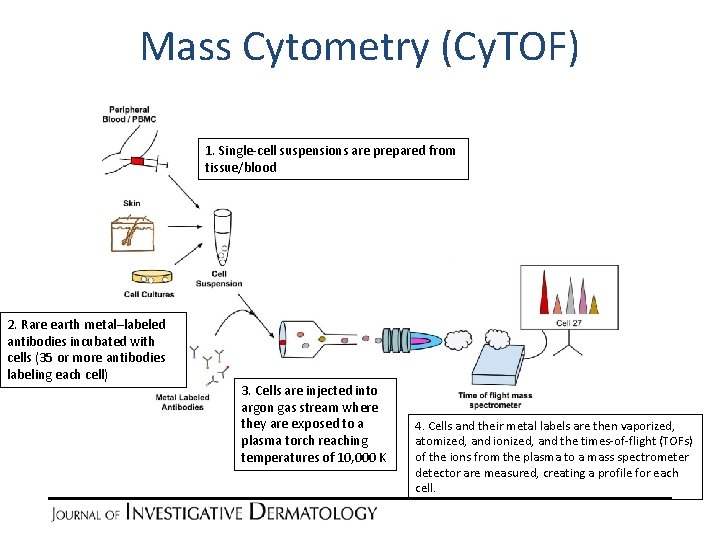
Mass Cytometry (Cy. TOF) 1. Single-cell suspensions are prepared from tissue/blood 2. Rare earth metal–labeled antibodies incubated with cells (35 or more antibodies labeling each cell) 3. Cells are injected into argon gas stream where they are exposed to a plasma torch reaching temperatures of 10, 000 K 4. Cells and their metal labels are then vaporized, atomized, and ionized, and the times-of-flight (TOFs) of the ions from the plasma to a mass spectrometer detector are measured, creating a profile for each cell.

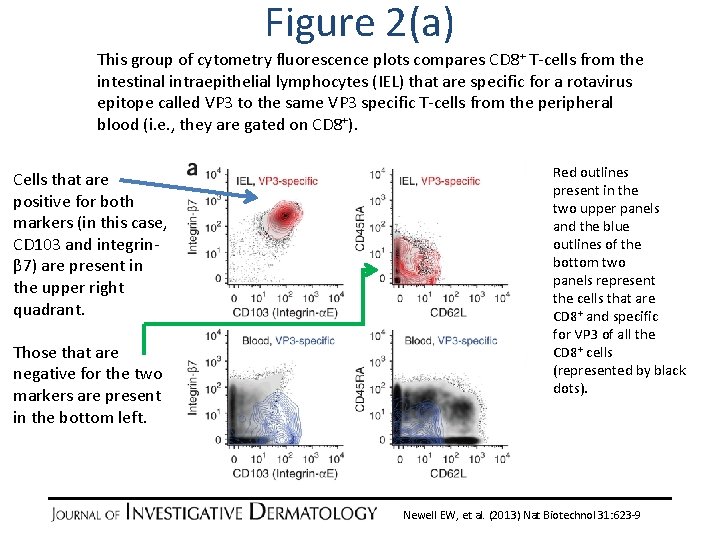
Figure 2(a) This group of cytometry fluorescence plots compares CD 8+ T-cells from the intestinal intraepithelial lymphocytes (IEL) that are specific for a rotavirus epitope called VP 3 to the same VP 3 specific T-cells from the peripheral blood (i. e. , they are gated on CD 8+). Cells that are positive for both markers (in this case, CD 103 and integrinβ 7) are present in the upper right quadrant. Those that are negative for the two markers are present in the bottom left. Red outlines present in the two upper panels and the blue outlines of the bottom two panels represent the cells that are CD 8+ and specific for VP 3 of all the CD 8+ cells (represented by black dots). Newell EW, et al. (2013) Nat Biotechnol 31: 623 -9

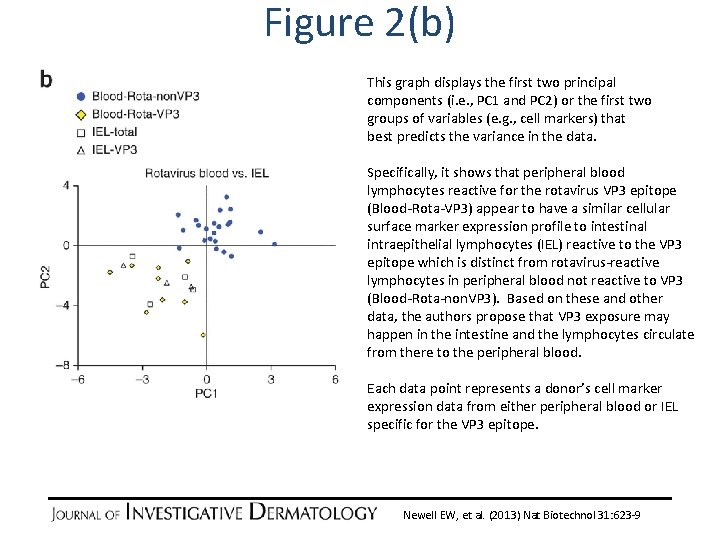
Figure 2(b) This graph displays the first two principal components (i. e. , PC 1 and PC 2) or the first two groups of variables (e. g. , cell markers) that best predicts the variance in the data. Specifically, it shows that peripheral blood lymphocytes reactive for the rotavirus VP 3 epitope (Blood-Rota-VP 3) appear to have a similar cellular surface marker expression profile to intestinal intraepithelial lymphocytes (IEL) reactive to the VP 3 epitope which is distinct from rotavirus-reactive lymphocytes in peripheral blood not reactive to VP 3 (Blood-Rota-non. VP 3). Based on these and other data, the authors propose that VP 3 exposure may happen in the intestine and the lymphocytes circulate from there to the peripheral blood. Each data point represents a donor’s cell marker expression data from either peripheral blood or IEL specific for the VP 3 epitope. Newell EW, et al. (2013) Nat Biotechnol 31: 623 -9

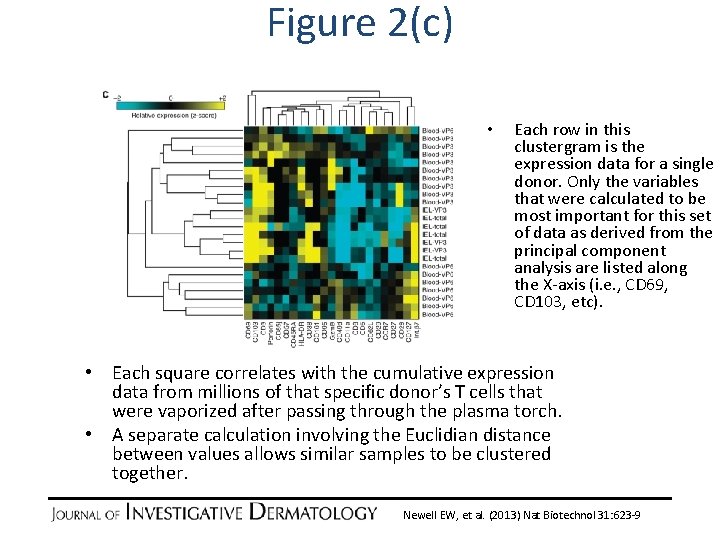
Figure 2(c) • Each row in this clustergram is the expression data for a single donor. Only the variables that were calculated to be most important for this set of data as derived from the principal component analysis are listed along the X-axis (i. e. , CD 69, CD 103, etc). • Each square correlates with the cumulative expression data from millions of that specific donor’s T cells that were vaporized after passing through the plasma torch. • A separate calculation involving the Euclidian distance between values allows similar samples to be clustered together. Newell EW, et al. (2013) Nat Biotechnol 31: 623 -9

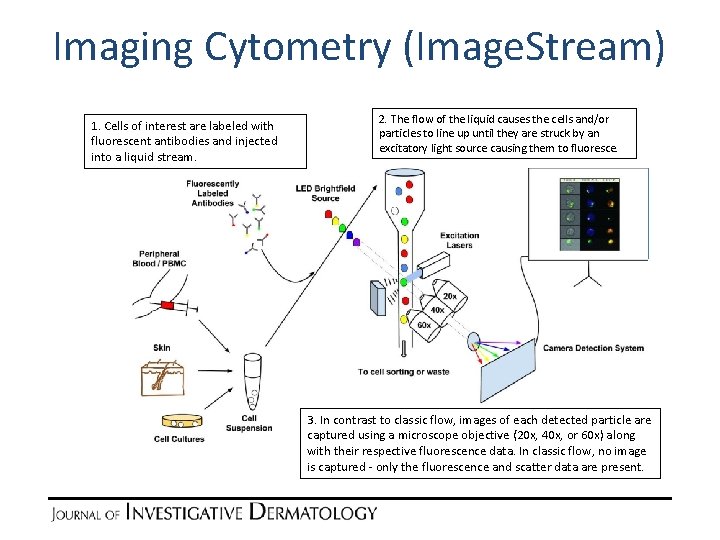
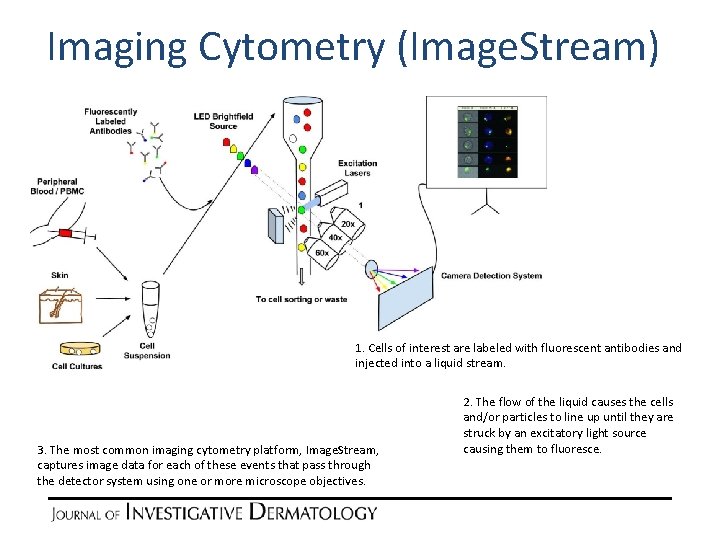
Imaging Cytometry (Image. Stream) 1. Cells of interest are labeled with fluorescent antibodies and injected into a liquid stream. 2. The flow of the liquid causes the cells and/or particles to line up until they are struck by an excitatory light source causing them to fluoresce. 3. In contrast to classic flow, images of each detected particle are captured using a microscope objective (20 x, 40 x, or 60 x) along with their respective fluorescence data. In classic flow, no image is captured - only the fluorescence and scatter data are present.

Imaging Cytometry (Image. Stream) 1. Cells of interest are labeled with fluorescent antibodies and injected into a liquid stream. 3. The most common imaging cytometry platform, Image. Stream, captures image data for each of these events that pass through the detector system using one or more microscope objectives. 2. The flow of the liquid causes the cells and/or particles to line up until they are struck by an excitatory light source causing them to fluoresce.

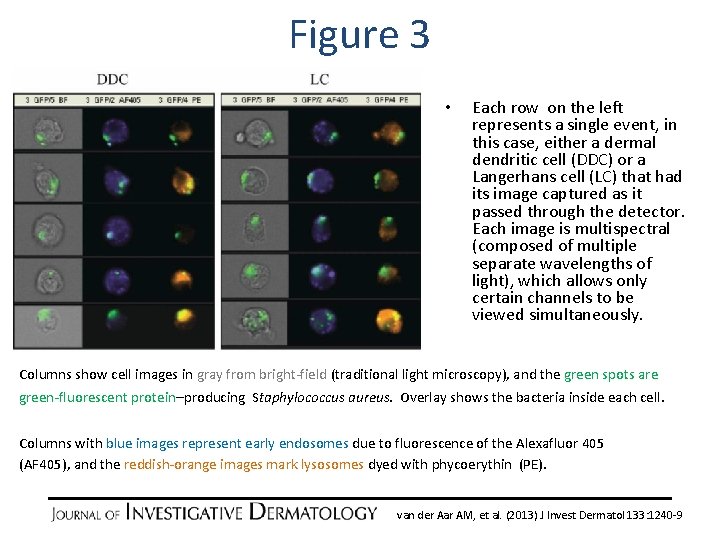
Figure 3 • Each row on the left represents a single event, in this case, either a dermal dendritic cell (DDC) or a Langerhans cell (LC) that had its image captured as it passed through the detector. Each image is multispectral (composed of multiple separate wavelengths of light), which allows only certain channels to be viewed simultaneously. Columns show cell images in gray from bright-field (traditional light microscopy), and the green spots are green-fluorescent protein–producing Staphylococcus aureus. Overlay shows the bacteria inside each cell. Columns with blue images represent early endosomes due to fluorescence of the Alexafluor 405 (AF 405), and the reddish-orange images mark lysosomes dyed with phycoerythin (PE). van der Aar AM, et al. (2013) J Invest Dermatol 133: 1240 -9

SUMMARY • Mass cytometry (Cy. TOF) and imaging cytometry (Image. Stream) are technological offshoots of traditional flow cytometry that allow characterization of multiple simultaneous cellular and subcellular parameters. • Mass cytometry uses rare earth metal isotopes to label cells that are then read by a mass spectrometer, allowing for more than 35 labels on each cell. • Imaging cytometry (Image. Stream) combines high-resolution light and fluorescent microscopy to capture an image for each event passing through the detector. • During the mass cytometry (Cy. TOF) process the cells are destroyed, making subsequent cell sorting and analysis impossible. • Imaging cytometry (Image. Stream) suffers from fluorescent dye “spillover, ” which continues to limit its multiplexing capabilities. • The data analysis for both techniques can be complicated given the highly multiplexed nature of these techniques.