Representing the Immune Epitope Database in OWL Jason

Representing the Immune Epitope Database in OWL Jason A. Greenbaum 1, Randi Vita 1, Laura Zarebski 1, Hussein Emami 2, Alessandro Sette 1, Alan Ruttenberg 3, and Bjoern Peters 1 1 La Jolla Institute for Allergy and Immunology 2 Science Applications International Corporation 3 Science Commons

Overview • Background – Immune epitopes – Epitope mapping experiments – The Immune Epitope Database (IEDB) • IEDB development cycle – Ontology development – Database design – Content curation • Database export into OWL
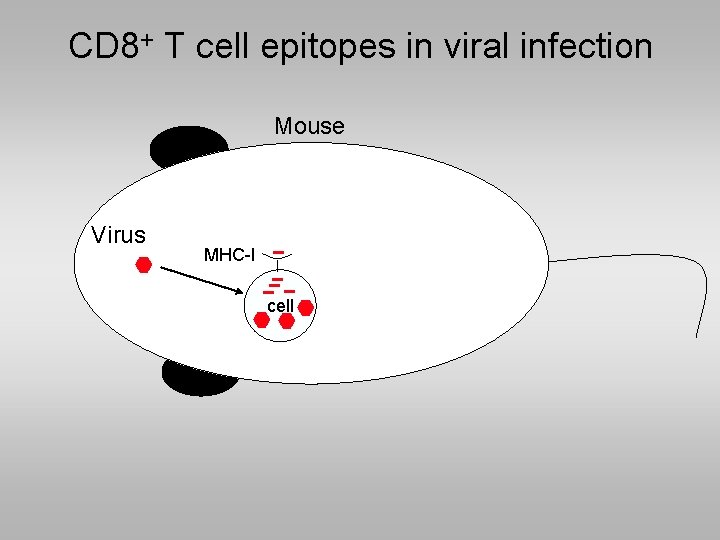
CD 8+ T cell epitopes in viral infection Mouse Virus MHC-I cell
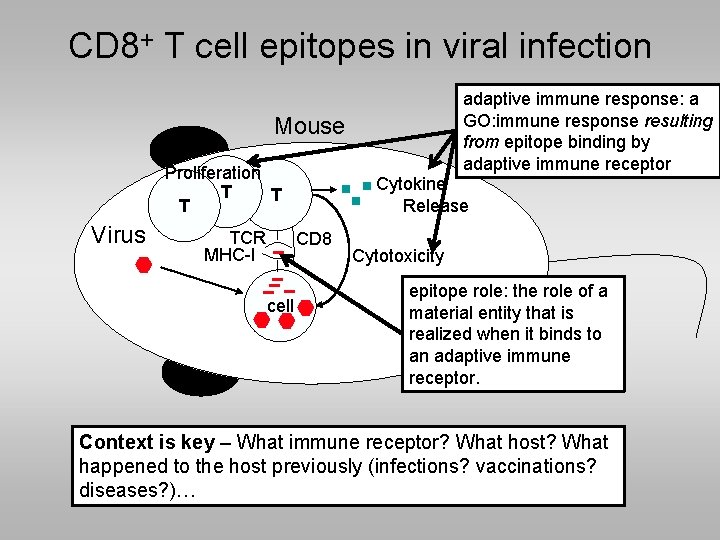
CD 8+ T cell epitopes in viral infection adaptive immune response: a GO: immune response resulting from epitope binding by adaptive immune receptor Mouse Proliferation T T T Virus TCR MHC-I Cytokine Release CD 8 cell Cytotoxicity epitope role: the role of a material entity that is realized when it binds to an adaptive immune receptor. Context is key – What immune receptor? What host? What happened to the host previously (infections? vaccinations? diseases? )…
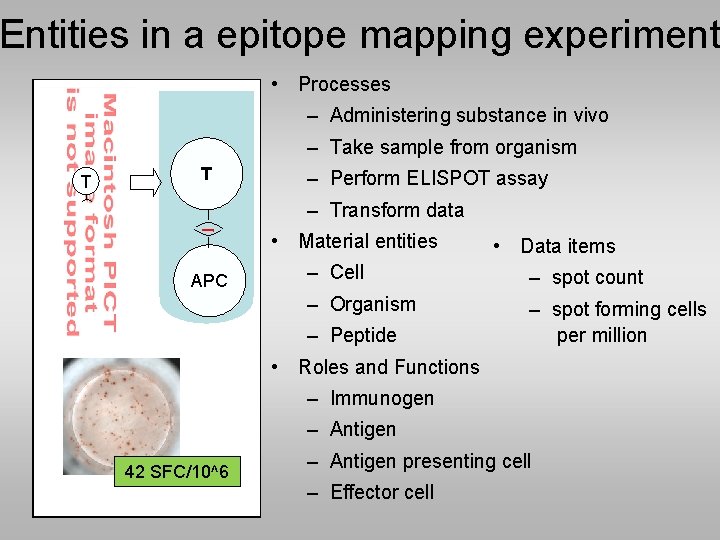
Entities in a epitope mapping experiment • Processes – Administering substance in vivo – Take sample from organism T T – Perform ELISPOT assay – Transform data • Material entities APC • Data items – Cell – spot count – Organism – spot forming cells per million – Peptide • Roles and Functions – Immunogen – Antigen 42 SFC/10^6 – Antigen presenting cell – Effector cell
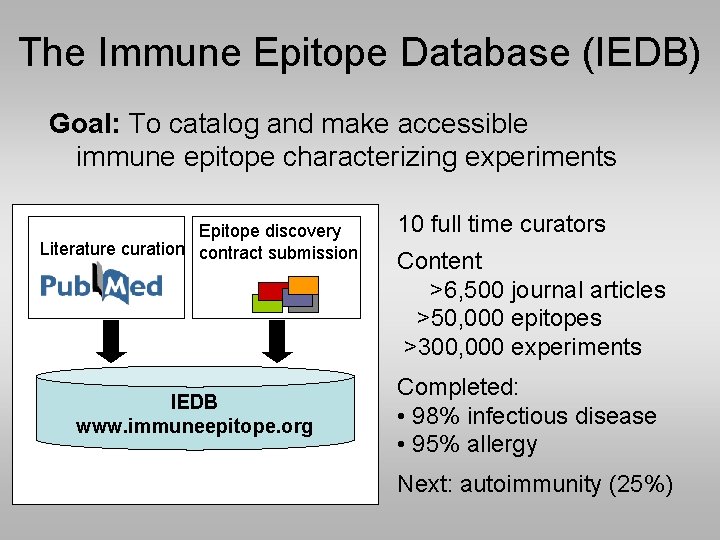
The Immune Epitope Database (IEDB) Goal: To catalog and make accessible immune epitope characterizing experiments Epitope discovery Literature curation contract submission IEDB www. immuneepitope. org 10 full time curators Content >6, 500 journal articles >50, 000 epitopes >300, 000 experiments Completed: • 98% infectious disease • 95% allergy Next: autoimmunity (25%)
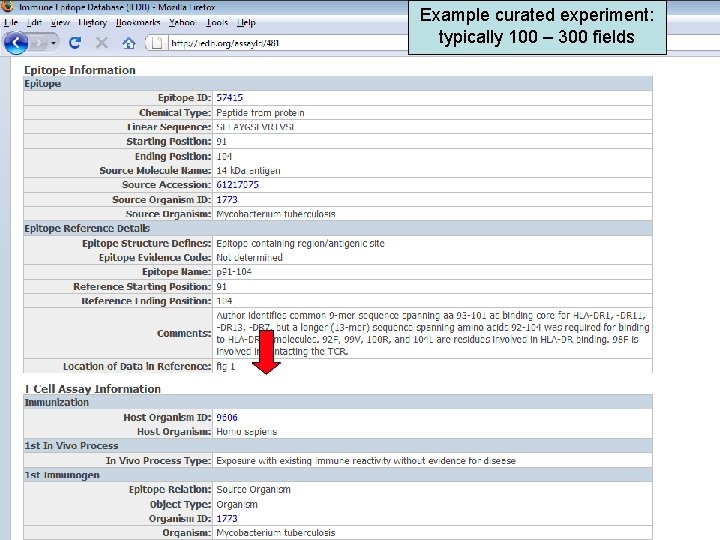
Example curated experiment: typically 100 – 300 fields
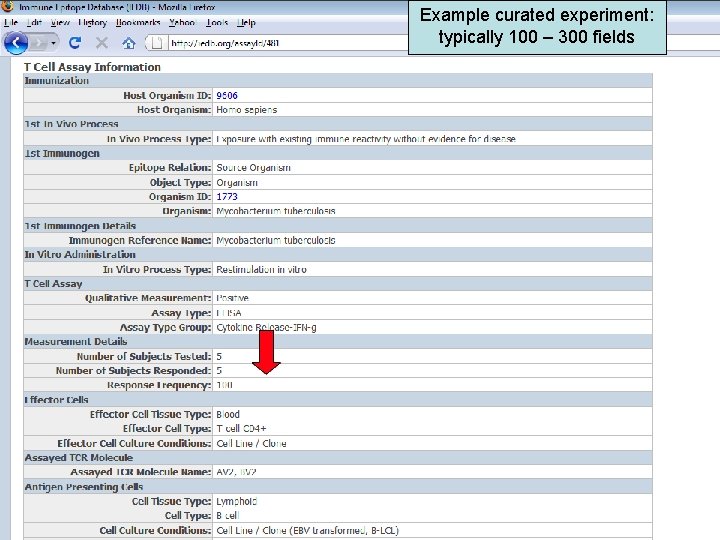
Example curated experiment: typically 100 – 300 fields
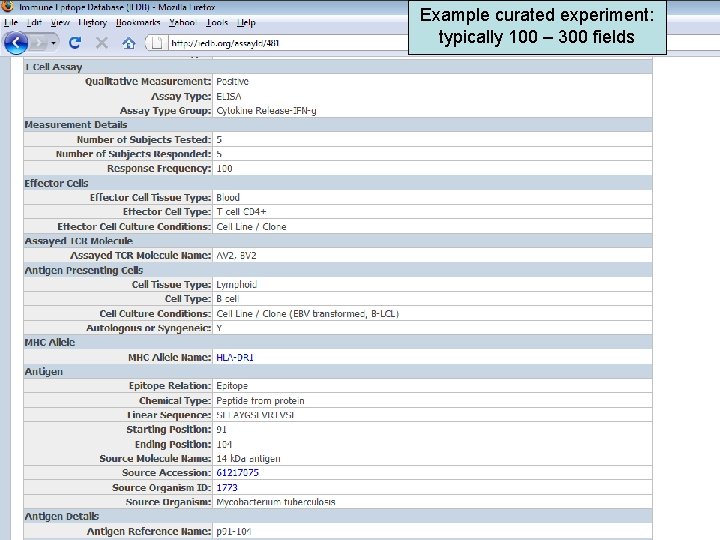
Example curated experiment: typically 100 – 300 fields

Summary I • Immune epitopes are the molecular entities recognized by adaptive immune receptors • The IEDB catalogs experiments defining immune epitopes Large amounts of complex data, which poses challenges for data consistency

Overview • Background – Immune epitopes – The Immune Epitope Database (IEDB) • IEDB development cycle – Ontology development – Database design – Content curation • Database export into OWL
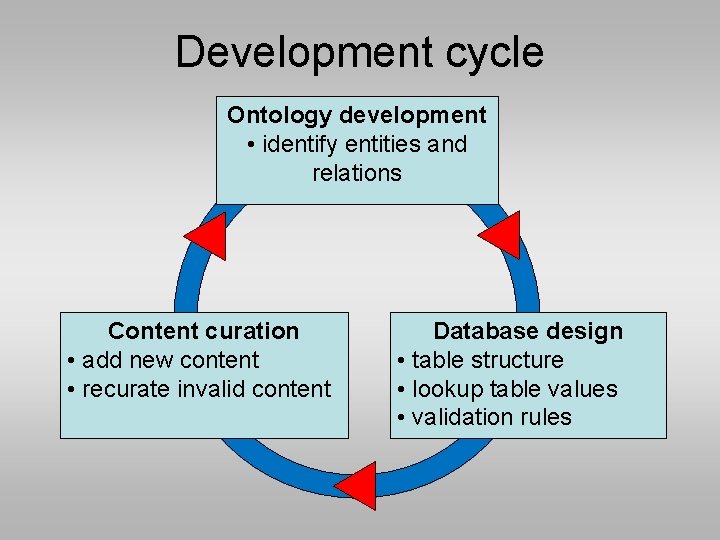
Development cycle Ontology development • identify entities and relations Content curation • add new content • recurate invalid content Database design • table structure • lookup table values • validation rules
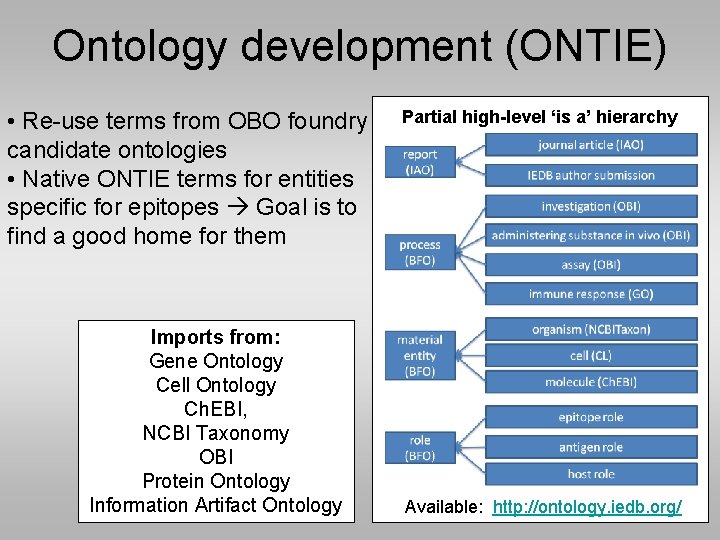
Ontology development (ONTIE) • Re-use terms from OBO foundry candidate ontologies • Native ONTIE terms for entities specific for epitopes Goal is to find a good home for them Imports from: Gene Ontology Cell Ontology Ch. EBI, NCBI Taxonomy OBI Protein Ontology Information Artifact Ontology Partial high-level ‘is a’ hierarchy Available: http: //ontology. iedb. org/
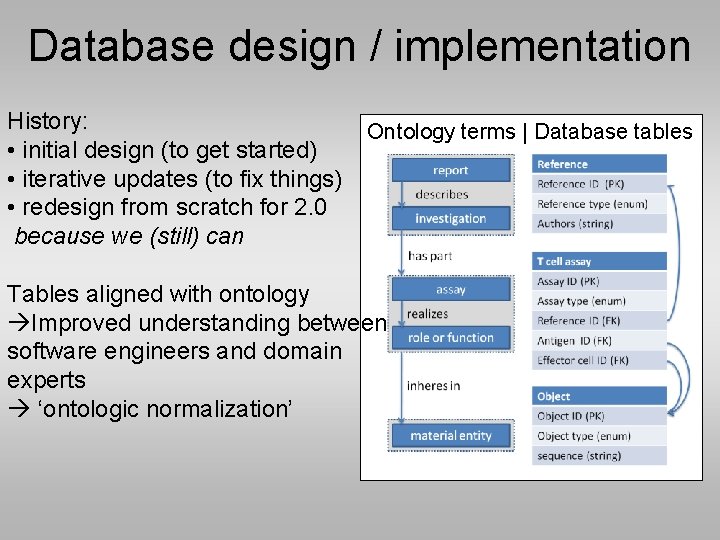
Database design / implementation History: • initial design (to get started) • iterative updates (to fix things) • redesign from scratch for 2. 0 because we (still) can Ontology terms | Database tables Tables aligned with ontology Improved understanding between software engineers and domain experts ‘ontologic normalization’
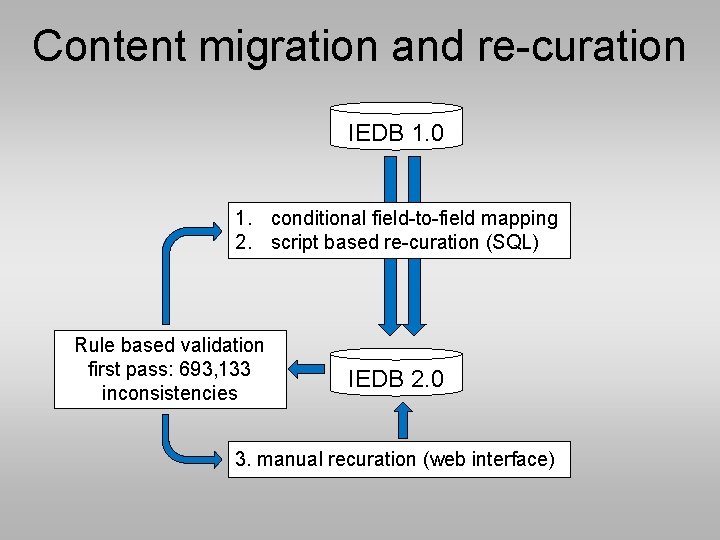
Content migration and re-curation IEDB 1. 0 1. conditional field-to-field mapping 2. script based re-curation (SQL) Rule based validation first pass: 693, 133 inconsistencies IEDB 2. 0 3. manual recuration (web interface)

Summary II • Application specific ontology (ONTIE) developed based on OBO foundry principles, and relying heavily on OBI • Database re-designed and structure aligned with the ontology • Data migrated and consistency enforced by rule based validation engine

Overview • Background – Immune epitopes – The Immune Epitope Database (IEDB) • IEDB development cycle – Ontology development – Database design – Content curation • Database export into OWL
Database export into OWL Subset of IEDB 2. 0
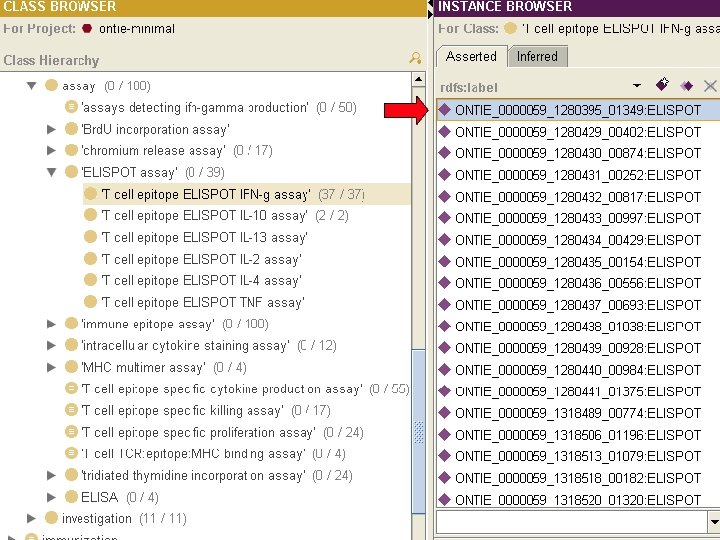
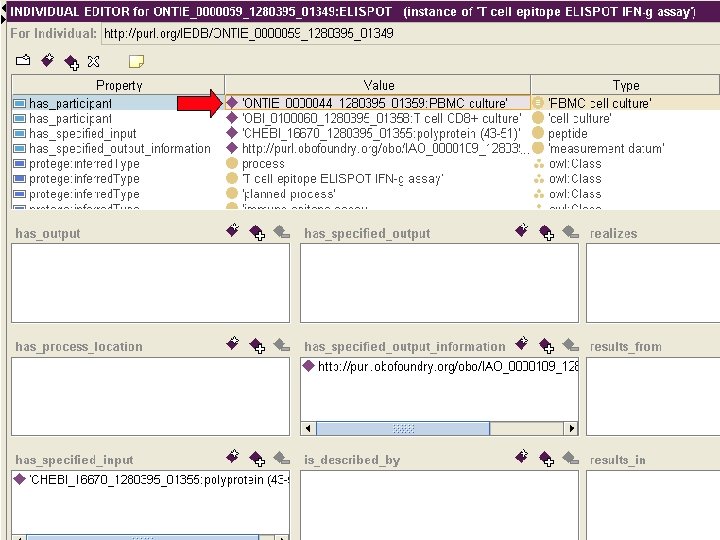
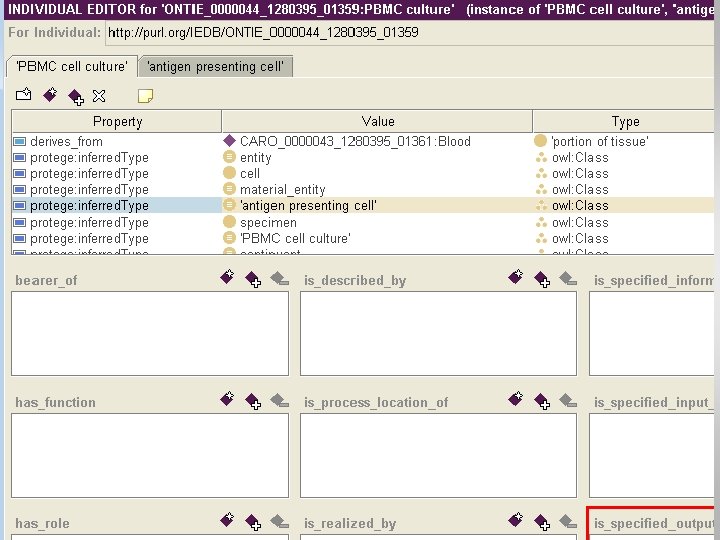

Advantages of OWL export • Allows to directly use ontology and OWL reasoner to perform consistency checks • Provides expressive query language within the IEDB • Enables query across integrated biomedical databases.

Future Work • Provide IEDB in triple store / access through SPARQL queries • Complete ontology development and OWL export for all data in the IEDB • Overcome technical challenges (Pellet takes 1 minute to classify 100 assays; 300, 000 in IEDB…) • Overcome ontological challenges (cells, peptides, negative data, …)

THANKS! OBI Consortium - http: //obi-ontology. org Alan Ruttenberg – Science Commons IEDB Team - www. iedb. org La Jolla Institute for Allergy & Immunology SAIC • Scott Stewart • Tom Carolan • Hussein Emami San Diego Supercomputer Center • Phil Bourne • Julia Ponomarenko • Zhanyang Zhu Technical University of Denmark • Ole Lund • Morten Nielsen University of Copenhagen • Søren Buus
- Slides: 24