Repetitive Beta Folds Form Function and Properties Overview

Repetitive Beta Folds Form, Function, and Properties

Overview of Presentation Introduction to traditional beta-helix fold – Structural and functional properties – Structure prediction by Beta. Wrap program Introduction to trimeric viral attachment fibers – Structural comparison to traditional beta-helices – Current computational approaches

Basic Parallel Beta-Helix Beta-helix is an all beta fold Three strands per rung Right-handed helix (RHBH) Also left-handed helices (LHBH) Three faces form a prism Function: Sugar cleavage Mainly occurs in bacterial pathogens King lab studies RHBH Tailspike

Analyzing Beta-Helices Solved structures – – RHBH: 5 SCOP Super. Families LHBH: 2 SCOP Super. Families 48 solved structures in PDB 8 HSSP representatives Predicting novel beta-helices – Homology modeling, threading, and HMMs do not successfully predict occurrence in cross-validation Beta. Wrap (King, Berger et al. 2001) successfully predicts RHBH

Lessons from Beta. Wrap Joint sequence-structure analysis important – Discovered conserved hairpin turn – Discovered internally packed asparagines Beta-strand packing interactions are important – Beta. Wrap energy function uses strand-to-strand packing probabilities Prediction is not enough – Beta. Wrap does not predict active site, etc. – Other methods (rotamer libraries etc. ) may supplement initial prediction

Trimeric Viral Attachment Fiber Proteins
Trimeric Viral Attachment Fibers King’s interest in beta helix led to interest in two new folds 1. Triple beta-helix (TBH) 2. Triple beta-spiral (TBS) 1. These two folds are our current research area – – – Consist of three identical interacting chains TBH is structurally similar to beta-helix TBS is structurally distinct Both folds characterized by unusual stability to heat, protease, and detergent

Triple Beta-Helix • Described by van Raaij et al. in JMB (2001) • Homo. Trimeric (consists of three identical chains) • Two solved structures – Portion of T 4 short tail fibre Swiss. Prot: P 10390 – Cell puncturing device of T 4 Swiss. Prot: P 16009
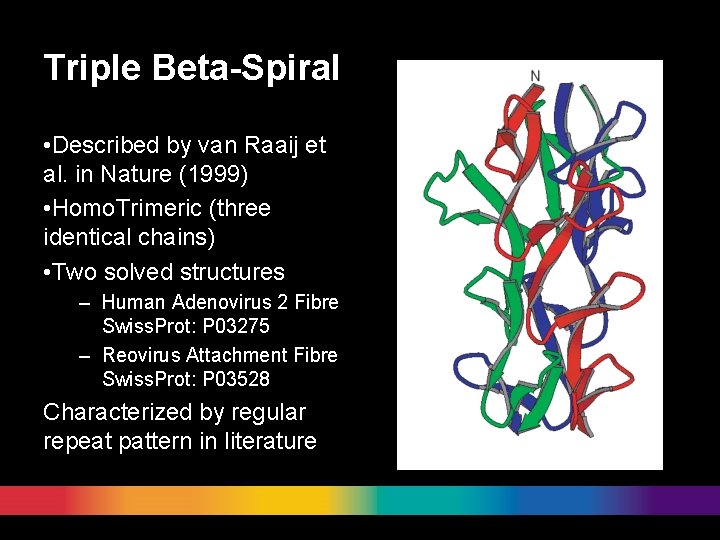
Triple Beta-Spiral • Described by van Raaij et al. in Nature (1999) • Homo. Trimeric (three identical chains) • Two solved structures – Human Adenovirus 2 Fibre Swiss. Prot: P 03275 – Reovirus Attachment Fibre Swiss. Prot: P 03528 Characterized by regular repeat pattern in literature
Preliminary Analysis TBS more regular than TBH – TBS characterized by sequence repeat – Can use standard regex techniques (like PROSITE) to find many putative TBSs – See http: //www. baobob. net/cgi-bin/repeat/stored-queries. pl TBH has so far defied basic characterization – Only two solved structures – The quasi-repeat is less regular than the TBS
Current Research What are we trying to do with TBH and TBS? – There are too few for rigorous prediction tool – Right now we are just “characterizing” them – Searching for sequence-structure patterns – Searching for unique properties – Searching for repetitive sequence motifs • Regular Expression is first attempt • Search with PSSM sequence profile
Repetitive Sequence Motif Search Existing Methods for repetitive motif search – RADAR (Heger & Holm) and others attempt this – Existing methods do not find the adeno repeat – TBH repeat is not regular enough to search Our approaches (tried so far…) – Basic regular expression (more in supplemental) – PSSM characterizing repeat (in progress)

Thank you Peter Weigele (pweigele@mit. edu) and Eben Scanlon (eben@mit. edu)

Supplemental Slides

TBS Information – Human Adenovirus 2 Fiber and Reovirus Attachment Protein σ1 have 27% sequence identity, 52% sequence similarity – Searching Swiss. Prot for the Adenovirus repeat (regex) pattern with more than 6 occurrences finds 3158 matches – Searching Swiss. Prot for the Reovirus repeat (regex) pattern finds 37578 matches – PDB ID’s are 1 kke, 1 qiu
TBH Information – The T 4 Short tail fiber TBH and the T 4 cell puncturing TBH have 32% sequence identity and 61% sequence similarity – There is no clear repeat pattern in TBH – Tried PSSM and HMM models with alignments derived from known repeat strands in TBH • Have not yet figured out a way to restrict to matches with a large number of recurrent repeats • Also may want to add a high non-affine gap penalty beyond a certain extension – PDB ID’s are 1 H 6 W, 1 K 28 – Need to use PQS (http: //pqs. ebi. ac. uk) to get trimer image
- Slides: 16