Repeats Professor Dina Sokol The Graduate Center of





![Longest Common Extension LP[i] = longest common prefix between a string and its ith Longest Common Extension LP[i] = longest common prefix between a string and its ith](https://slidetodoc.com/presentation_image/4853dda87d1a2f5736f82a8800a59bcc/image-10.jpg)
![v Formally, we know position k that maximizes k+LP[k]. if k+LP[k] > i, we v Formally, we know position k that maximizes k+LP[k]. if k+LP[k] > i, we](https://slidetodoc.com/presentation_image/4853dda87d1a2f5736f82a8800a59bcc/image-12.jpg)
![Consider three cases for ℓ = k+LP[k]-i: LP[k] k i i-k+1 1 S[i+ℓ] ℓ Consider three cases for ℓ = k+LP[k]-i: LP[k] k i i-k+1 1 S[i+ℓ] ℓ](https://slidetodoc.com/presentation_image/4853dda87d1a2f5736f82a8800a59bcc/image-13.jpg)





![Runs v Recall: a maximal run r[i…j] is a repeat with period p whose Runs v Recall: a maximal run r[i…j] is a repeat with period p whose](https://slidetodoc.com/presentation_image/4853dda87d1a2f5736f82a8800a59bcc/image-24.jpg)



![Available Tools v TRF – Tandem Repeats Finder [Benson] v ATRHunter [Wexler et al. Available Tools v TRF – Tandem Repeats Finder [Benson] v ATRHunter [Wexler et al.](https://slidetodoc.com/presentation_image/4853dda87d1a2f5736f82a8800a59bcc/image-34.jpg)











- Slides: 63

Repeats Professor Dina Sokol The Graduate Center of the City University of N. Y. (http: //www. sci. brooklyn. cuny. edu/~sokol/)

2 or more copies v

Maximal Runs v. A – – v. A run is also called: periodic string repetition tandem repeat tandem array run that occurs in a larger string is maximal if it cannot be extended to the right or left. e. g. aaabcxabcabcabcabaa

Problem Definition v Input: a string S over alphabet v Output: all squares that occur in S. After we show an algorithm for squares, we show it can be used to find all maximal runs.

Naive Algorithm v v v Consider all possible pairs (i, j) 1≤i‹j≤n Compare the suffixes si … sn to sj … sn If the length of the match is at least j-i characters, then there is a square beginning at location i with size 2(j-i).

Naïve Algorithm Si j i i n j n Sj 123456789 Example: x x x a b c d x x x i=6 j=10

Time Complexity of Naïve Algorithm v Consider all pairs (i, j): O(n 2) pairs v Compairing per pair v Overall: the substrings: O(n) O(n 3) time.

Main and Lorentz algorithm 1. Use Landau/Vishkin or KMP to reduce the matching from O(n) to constant time. 2. Reduce the number of pairs considered from O(n 2) to O(n log n). How?

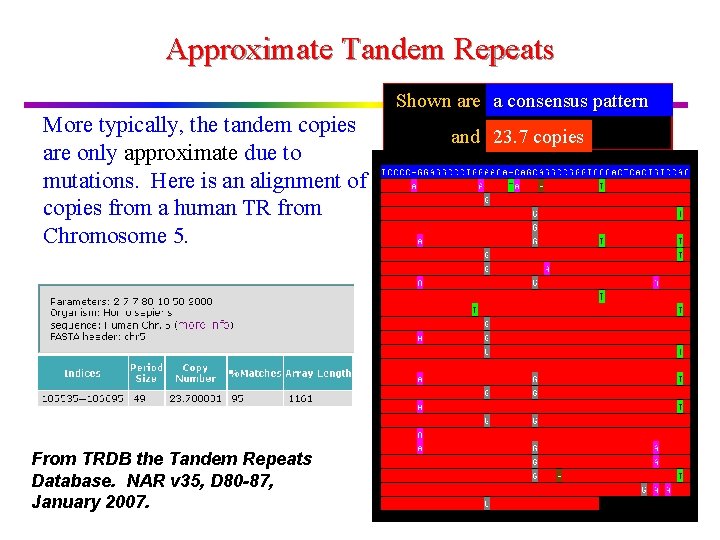
Longest Common Extension Function v
![Longest Common Extension LPi longest common prefix between a string and its ith Longest Common Extension LP[i] = longest common prefix between a string and its ith](https://slidetodoc.com/presentation_image/4853dda87d1a2f5736f82a8800a59bcc/image-10.jpg)
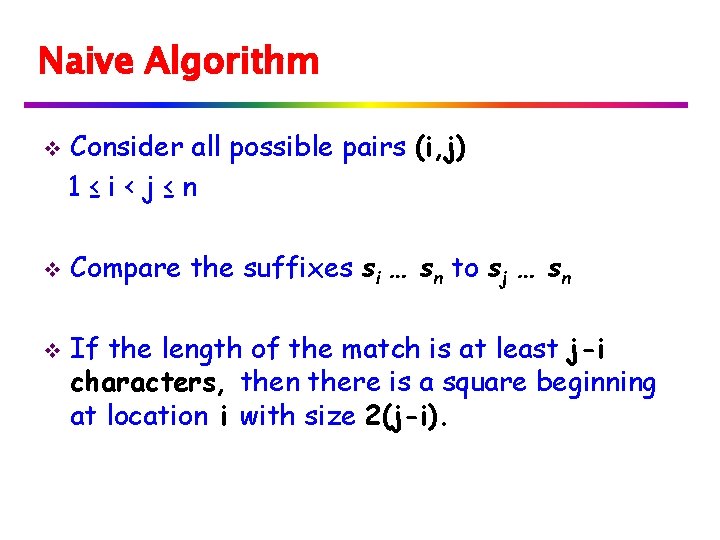
Longest Common Extension LP[i] = longest common prefix between a string and its ith suffix This can be computed using suffix trees with an LCP query between S and the ith suffix of S (constant time per location i).

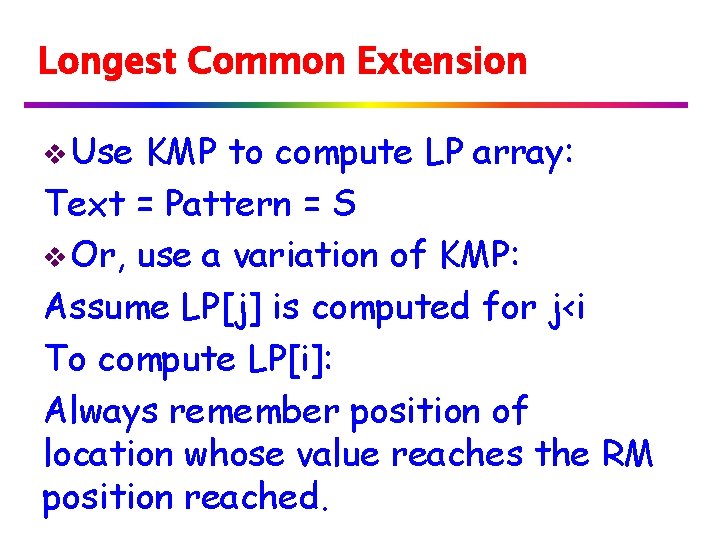
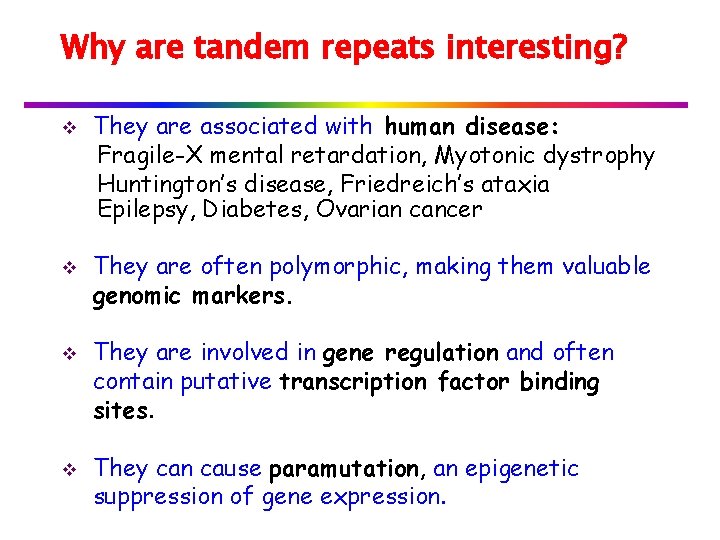
Longest Common Extension v Use KMP to compute LP array: Text = Pattern = S v Or, use a variation of KMP: Assume LP[j] is computed for j<i To compute LP[i]: Always remember position of location whose value reaches the RM position reached.
![v Formally we know position k that maximizes kLPk if kLPk i we v Formally, we know position k that maximizes k+LP[k]. if k+LP[k] > i, we](https://slidetodoc.com/presentation_image/4853dda87d1a2f5736f82a8800a59bcc/image-12.jpg)
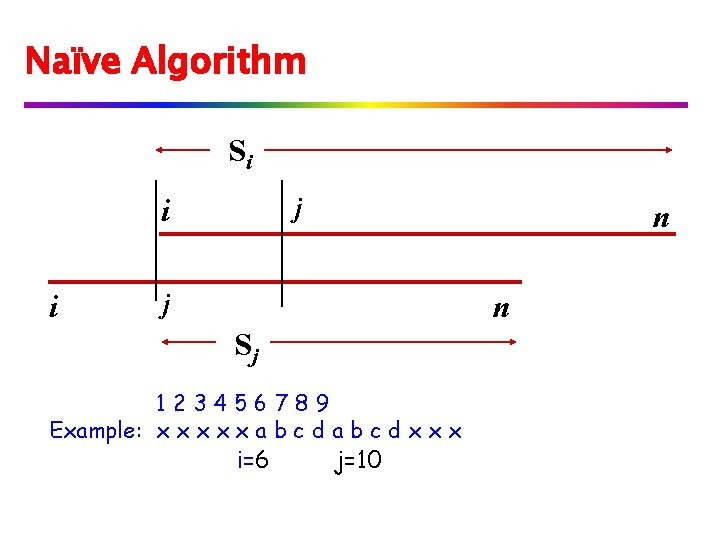
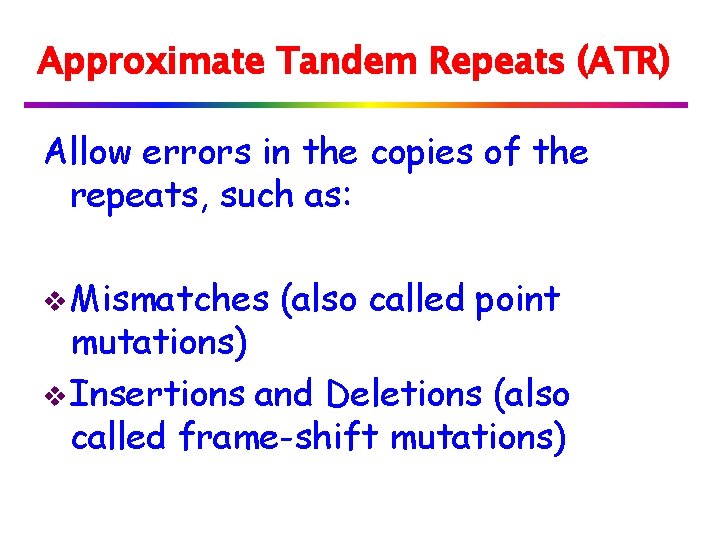
v Formally, we know position k that maximizes k+LP[k]. if k+LP[k] > i, we consider three cases for ℓ = k+LP[k]-i: if LP[i-k+1] > ℓ then LP[i] = ℓ if LP[i-k+1] < ℓ then LP[i]=LP[i-k+1] if LP[i-k+1] = ℓ then we continue comparing and position i becomes the new k.
![Consider three cases for ℓ kLPki LPk k i ik1 1 Siℓ ℓ Consider three cases for ℓ = k+LP[k]-i: LP[k] k i i-k+1 1 S[i+ℓ] ℓ](https://slidetodoc.com/presentation_image/4853dda87d1a2f5736f82a8800a59bcc/image-13.jpg)
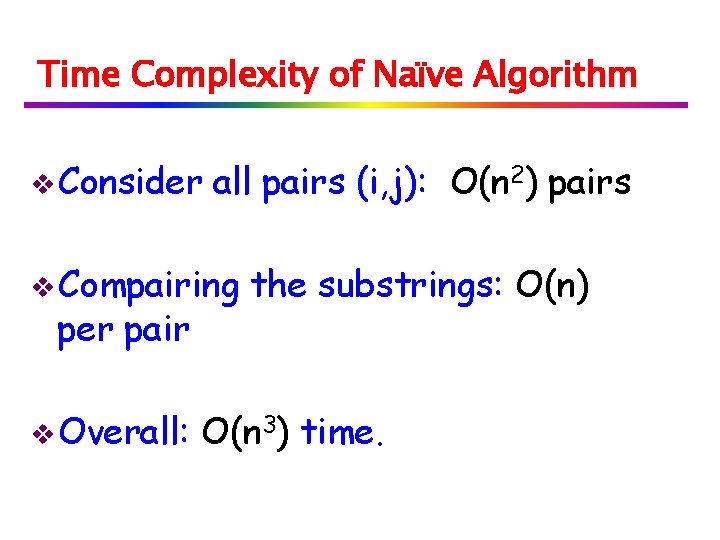
Consider three cases for ℓ = k+LP[k]-i: LP[k] k i i-k+1 1 S[i+ℓ] ℓ S[k+LP[k]] S[ℓ+1] if LP[i-k+1] (grey box) > ℓ then LP[i] = ℓ if LP[i-k+1] < ℓ then LP[i]=LP[i-k+1] if LP[i-k+1] = ℓ then we continue comparing and position i becomes the new k.

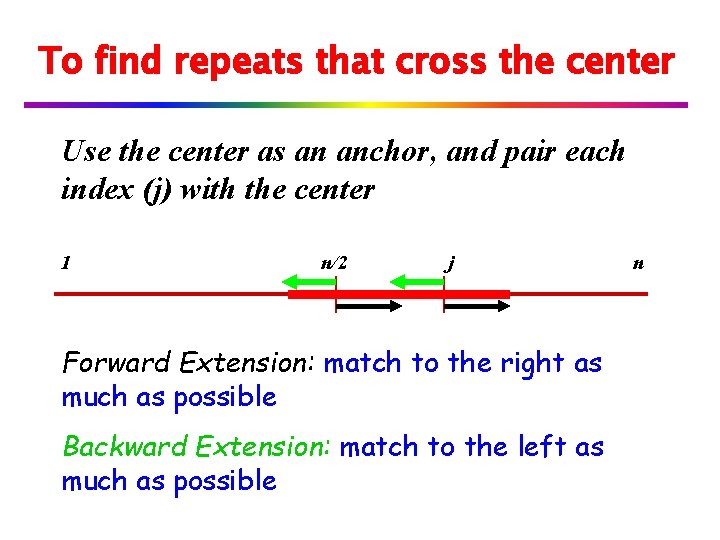
Find Repeats Crossing a Boundary v Find all repeats whose right half crosses the boundary (right repeats) v Find all repeats whose left half crosses the boundary (left repeats)

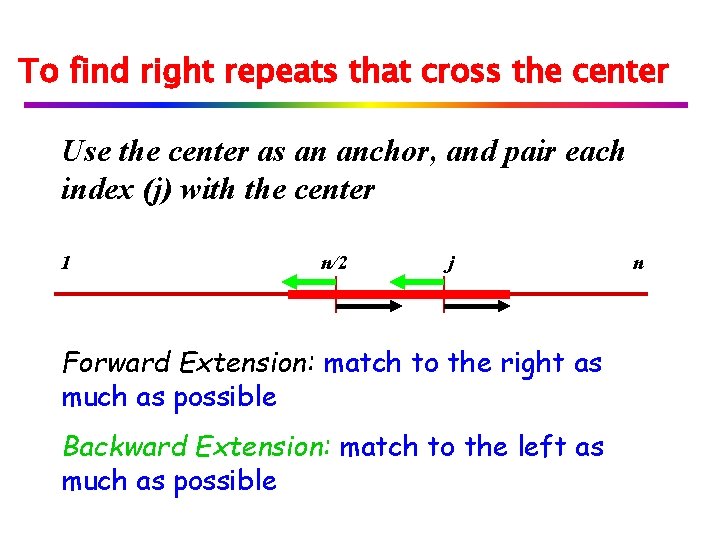
To find right repeats that cross the center Use the center as an anchor, and pair each index (j) with the center 1 n/2 j Forward Extension: match to the right as much as possible Backward Extension: match to the left as much as possible n

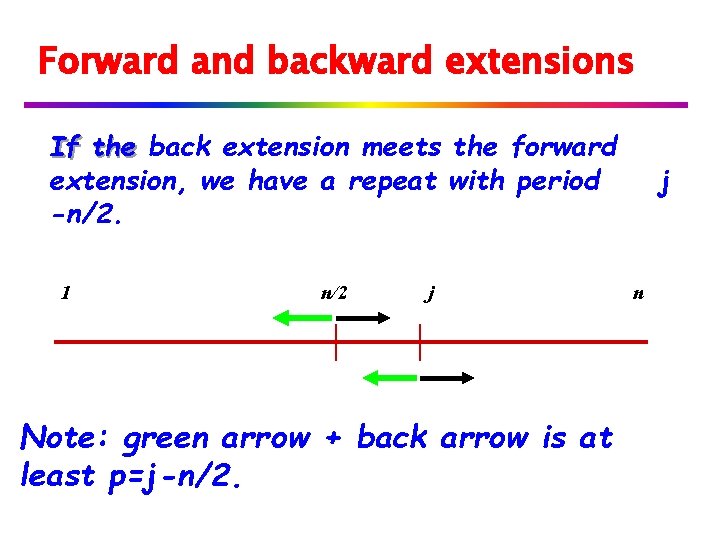
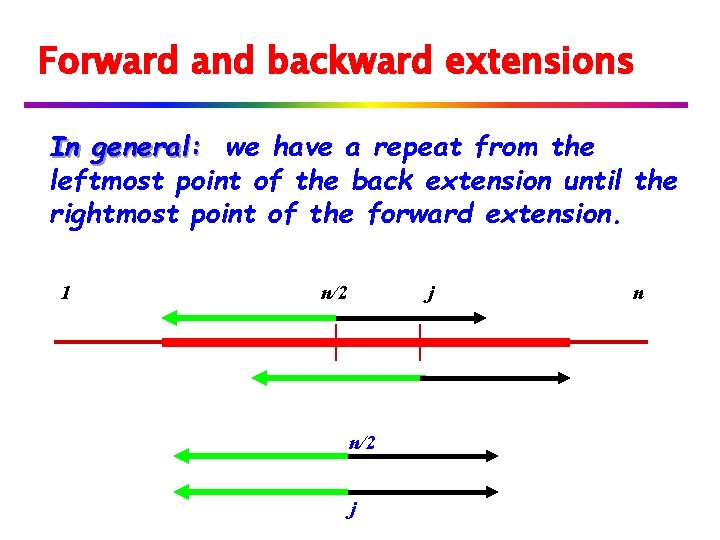
Forward and backward extensions If the back extension meets the forward extension, we have a repeat with period -n/2. 1 n/2 j Note: green arrow + back arrow is at least p=j-n/2. j n

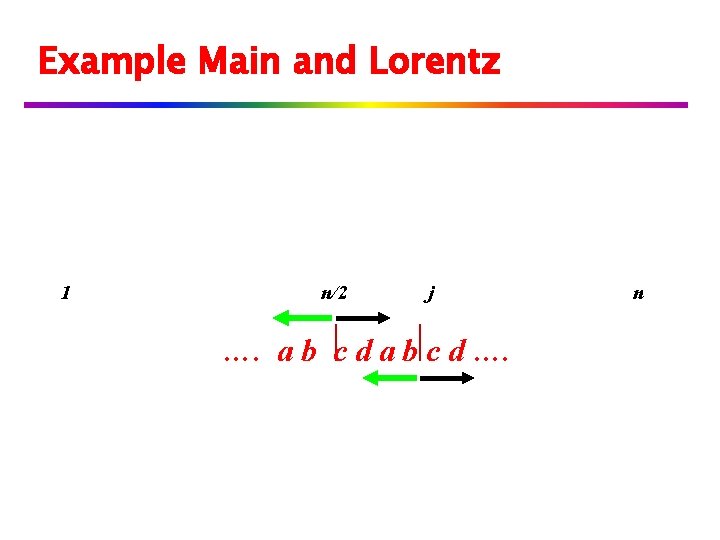
Example Main and Lorentz 1 n/2 j …. a b c d …. n

ALGOROTHM Find Right Repeats Right-Repeats(x, y) 1 LPy ← Longest-Prefix-Extension(y) 2 LSx|y← Longest-Suffix-Extension(x, y) 3 R←∅ 4 for p ← 1 to |y| do 5 if LSx|y(p) + LPy(p + 1) ≥ p then 6 r ← (m− LSx|y(p) + 1, m + p + LPy(p + 1)) 7 R←R∪{r} 8 return R

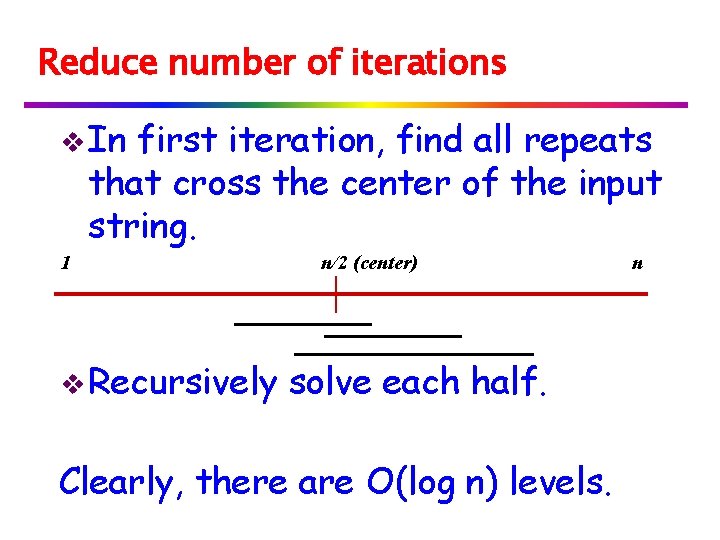
Reduce number of iterations v In first iteration, find all repeats that cross the center of the input string. 1 v Recursively n/2 (center) solve each half. Clearly, there are O(log n) levels. n

Overall Time: O(n log n) v Each iteration is done in linear time: every j along string pairs with the center in two directions, and longest common extension is done in constant time per j. v There are log n iterations.

Approximate Repeats v Suppose we have exactly two copies (square), and errors are introduced. v Assume that a Hamming Distance of k is allowed between the first and second copy of each repeat.

Use Kangaroo Jumps v Using the same framework of Main and Lorenz, pair every j with the center v Instead of computing the Longest Common Extension, use the Kangaroo method to find the positions of the first k mimsatches.

Landau-Schmidt-Sokol v
![Runs v Recall a maximal run rij is a repeat with period p whose Runs v Recall: a maximal run r[i…j] is a repeat with period p whose](https://slidetodoc.com/presentation_image/4853dda87d1a2f5736f82a8800a59bcc/image-24.jpg)
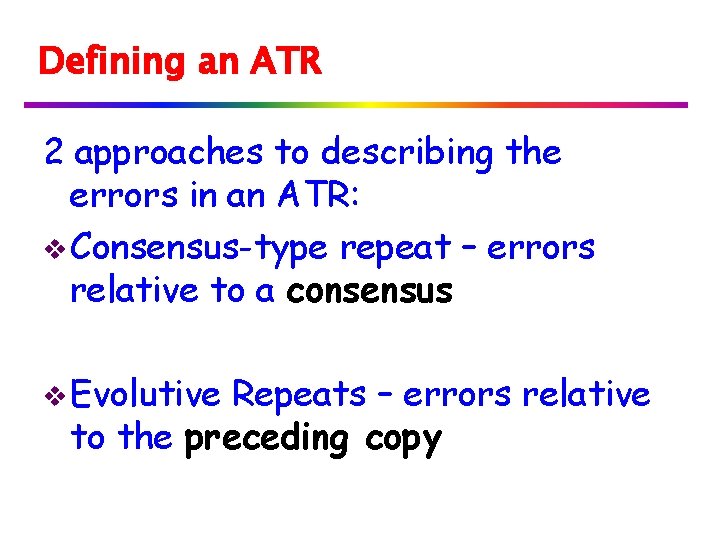
Runs v Recall: a maximal run r[i…j] is a repeat with period p whose length is at least 2 p, and it cannot be extended to the right or left. v the rational number (j-i+1)/p is the exponent of the run. example: alfalfa = (alf)7/3

Maximal Runs v Goal: Find all maximal runs in an input string S of length n. v Main and Lorentz can be trivially extended to find all maximal runs!

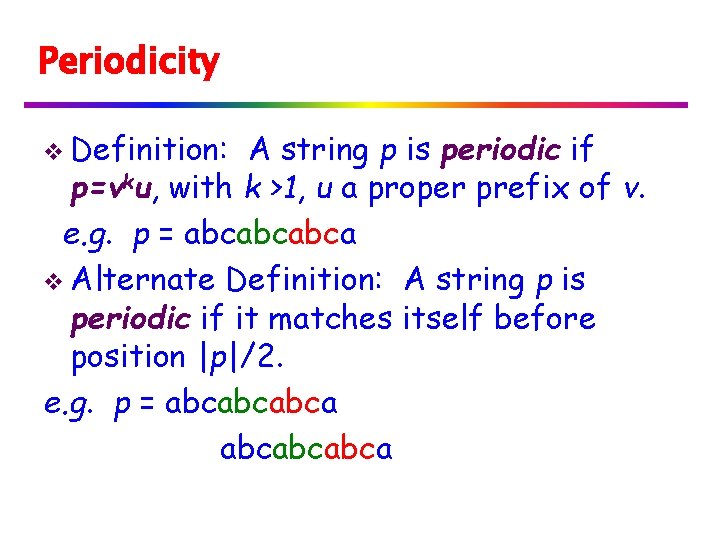
Periodicity v Definition: A string p is periodic if p=vku, with k >1, u a proper prefix of v. e. g. p = abcabcabca v Alternate Definition: A string p is periodic if it matches itself before position |p|/2. e. g. p = abcabcabca

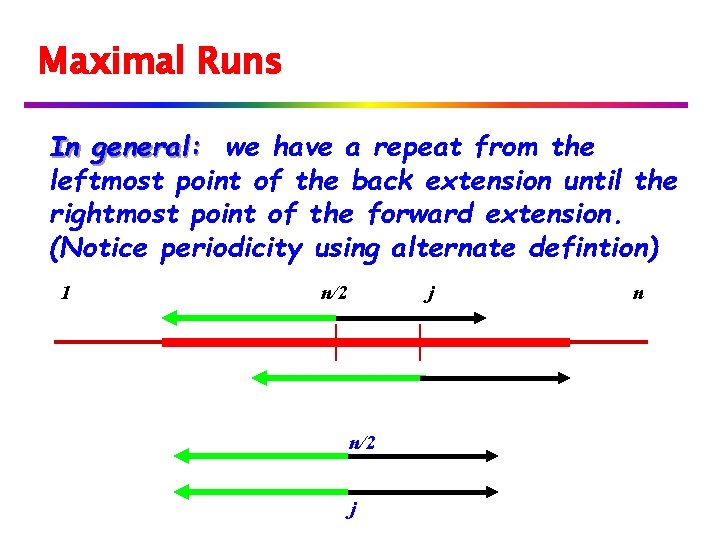
Maximal Runs In general: we have a repeat from the leftmost point of the back extension until the rightmost point of the forward extension. (Notice periodicity using alternate defintion) 1 n/2 j n

Approximate Runs v also called tandem repeats v need a distance measure v need a way to count errors – consider Hamming Distance measure – how to count mismatches v give 2 ideas (LSS and ACLS) v time permitting: continue to edit distance

What is a Tandem Repeat? A tandem repeat is a pattern of nucleotides that occurs consecutively 2 or more times. Example: The pattern CGT is repeated 5 times. …tcatacgt cgt cgttacaaacgtcttccgt…

Approximate Tandem Repeats More typically, the tandem copies are only approximate due to mutations. Here is an alignment of copies from a human TR from Chromosome 5. From TRDB the Tandem Repeats Database. NAR v 35, D 80 -87, January 2007. Shown are a consensus pattern and 23. 7 copies

Why are tandem repeats interesting? v v They are associated with human disease: Fragile-X mental retardation, Myotonic dystrophy Huntington’s disease, Friedreich’s ataxia Epilepsy, Diabetes, Ovarian cancer They are often polymorphic, making them valuable genomic markers. They are involved in gene regulation and often contain putative transcription factor binding sites. They can cause paramutation, an epigenetic suppression of gene expression.

Approximate Tandem Repeats (ATR) Allow errors in the copies of the repeats, such as: v Mismatches (also called point mutations) v Insertions and Deletions (also called frame-shift mutations)

Defining an ATR 2 approaches to describing the errors in an ATR: v Consensus-type repeat – errors relative to a consensus v Evolutive Repeats – errors relative to the preceding copy
![Available Tools v TRF Tandem Repeats Finder Benson v ATRHunter Wexler et al Available Tools v TRF – Tandem Repeats Finder [Benson] v ATRHunter [Wexler et al.](https://slidetodoc.com/presentation_image/4853dda87d1a2f5736f82a8800a59bcc/image-34.jpg)
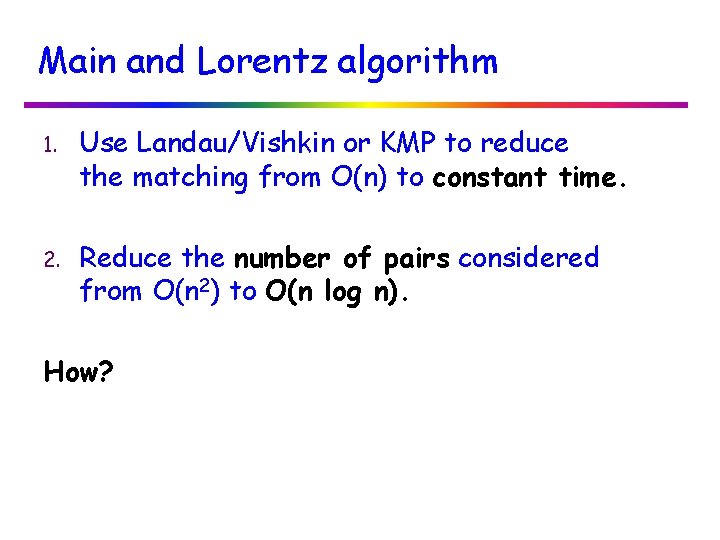
Available Tools v TRF – Tandem Repeats Finder [Benson] v ATRHunter [Wexler et al. ] v Tandem. SWAN [Boeva et al. ] - use heuristics and statistical methods. v mreps [Kolpakov and Kucherov] v [Landau/Schmidt/Sokol] - exhaustive search, allow only mismatches

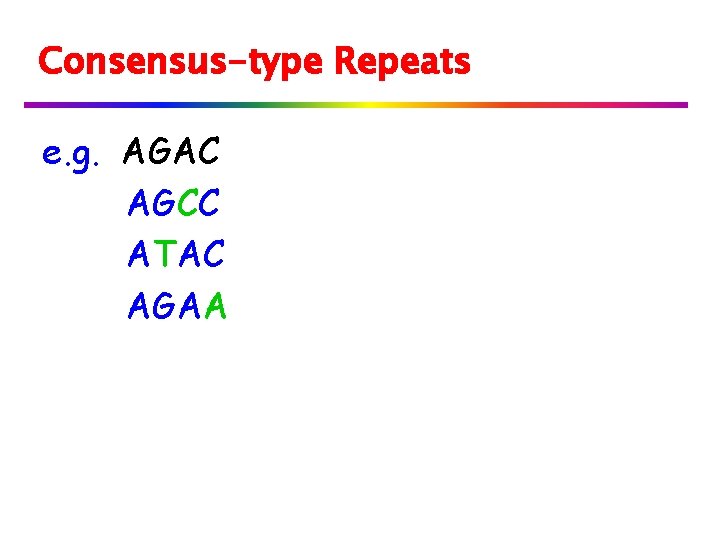
Consensus-type Repeats e. g. AGAC AGCC ATAC AGAA

Evolutive Tandem Repeats The assumption here is that each copy is derived from a neighbor copy. e. g. AGCC ACCT GCCT

Observation: Every consensus type repeat with k errors, is also an evolutive repeat with no more than 2 k errors. e. g. AGAC AGCC ATAC AGAA AGAC k = 3 consensus k = 6 evolutive

Our Goal v Perform an exhaustive search for all evolutive tandem repeats in a given sequence. v Allow up to k insertions, deletions, and mismatches.

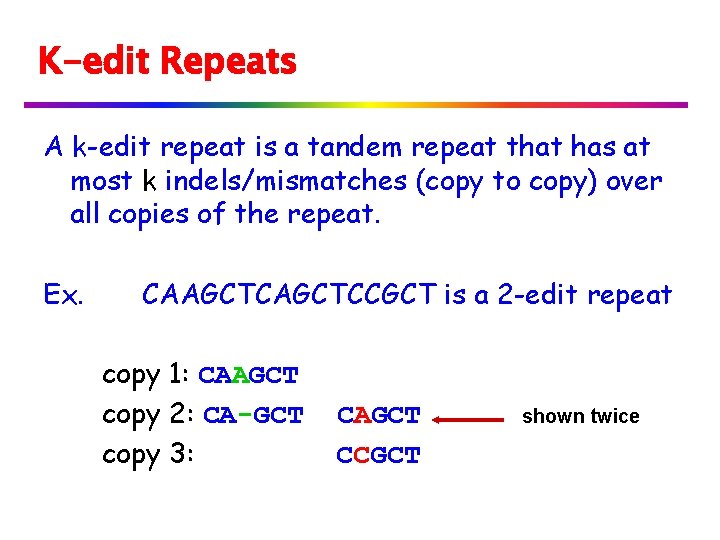
K-edit Repeats A k-edit repeat is a tandem repeat that has at most k indels/mismatches (copy to copy) over all copies of the repeat. Ex. CAAGCTCCGCT is a 2 -edit repeat copy 1: CAAGCT copy 2: CA-GCT copy 3: CAGCT CCGCT shown twice

Observation A string is a k-edit repeat iff there exists an alignment of the string with a proper suffix of itself, with ≤ k errors.

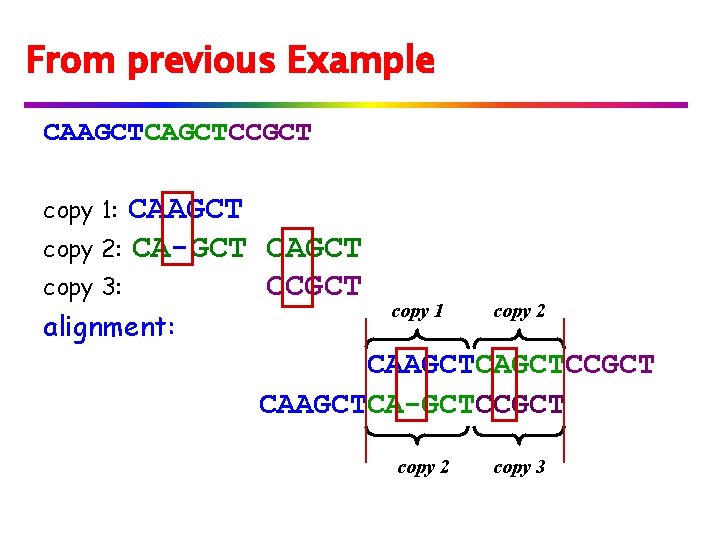
From previous Example CAAGCTCCGCT CAAGCT copy 2: CA-GCT CAGCT copy 3: CCGCT copy 1: alignment: copy 1 copy 2 CAAGCTCCGCT CAAGCTCA-GCTCCGCT copy 2 copy 3

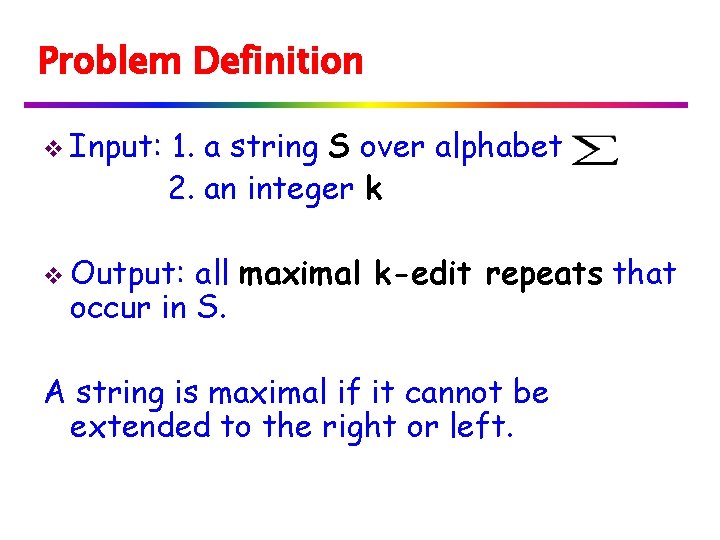
Problem Definition v Input: 1. a string S over alphabet 2. an integer k v Output: all maximal k-edit repeats that occur in S. A string is maximal if it cannot be extended to the right or left.

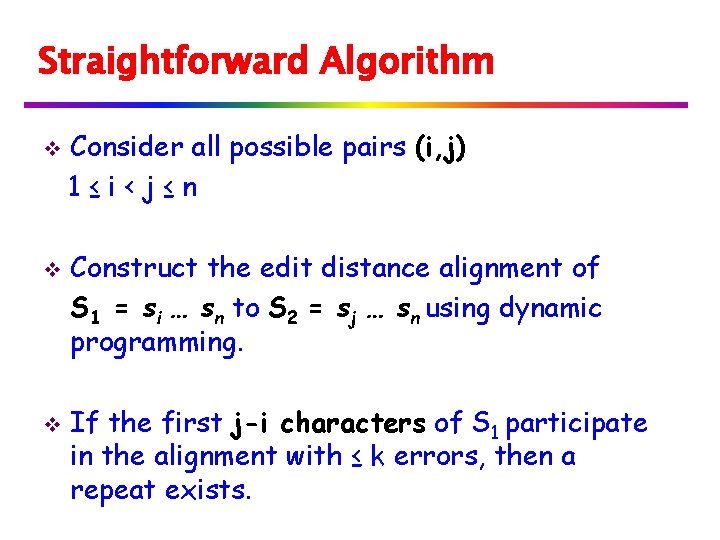
Straightforward Algorithm v v v Consider all possible pairs (i, j) 1≤i‹j≤n Construct the edit distance alignment of S 1 = si … sn to S 2 = sj … sn using dynamic programming. If the first j-i characters of S 1 participate in the alignment with ≤ k errors, then a repeat exists.

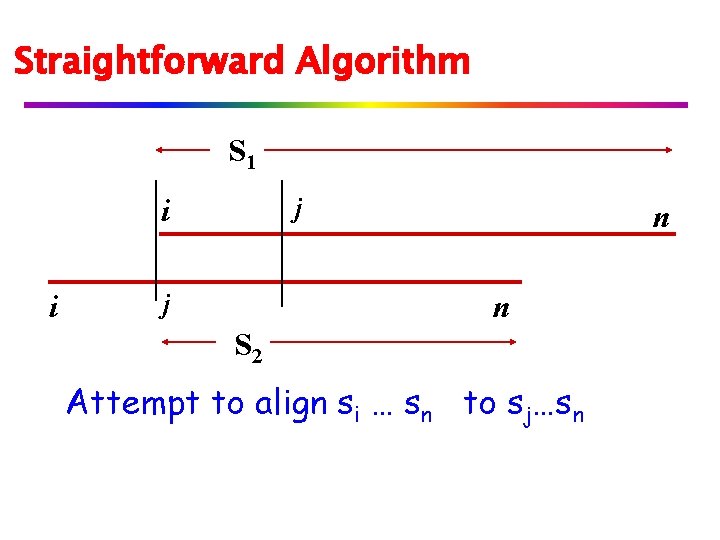
Straightforward Algorithm S 1 j i i j n n S 2 Attempt to align si … sn to sj…sn

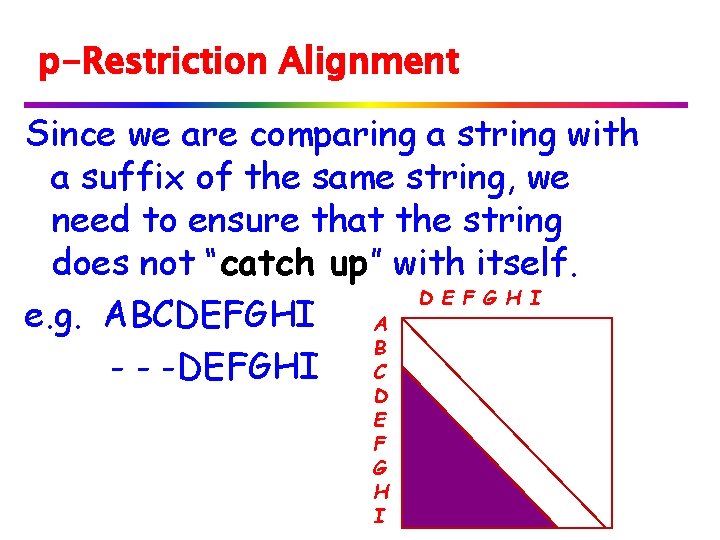
p-Restriction Alignment Since we are comparing a string with a suffix of the same string, we need to ensure that the string does not “catch up” with itself. D E F G H I e. g. ABCDEFGHI A B - - -DEFGHI C D E F G H I

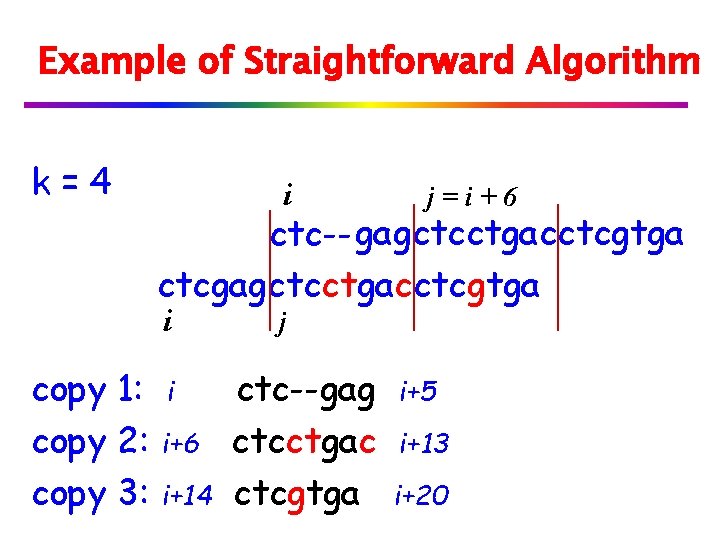
Example of Straightforward Algorithm k=4 i j=i+6 ctc-- gagctcctgacctcgtga ctcgagctcctgacctcgtga i copy 1: copy 2: copy 3: i i+6 i+14 j ctc--gag ctcctgac ctcgtga i+5 i+13 i+20

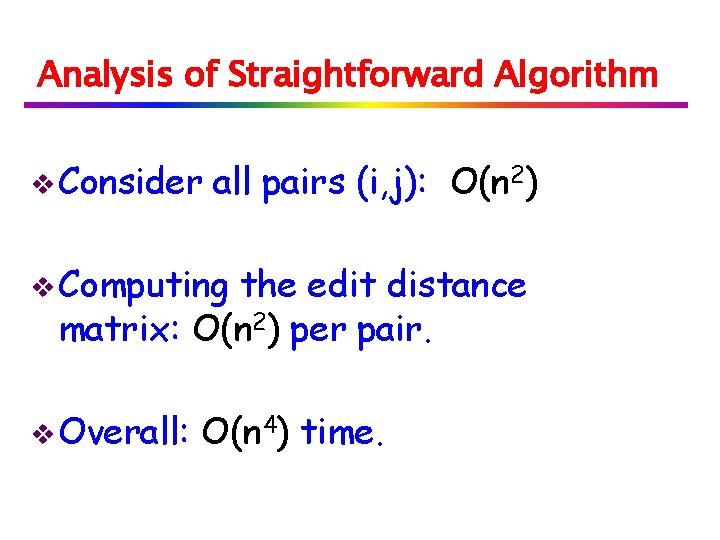
Analysis of Straightforward Algorithm v Consider all pairs (i, j): O(n 2) v Computing the edit distance matrix: O(n 2) per pair. v Overall: O(n 4) time.

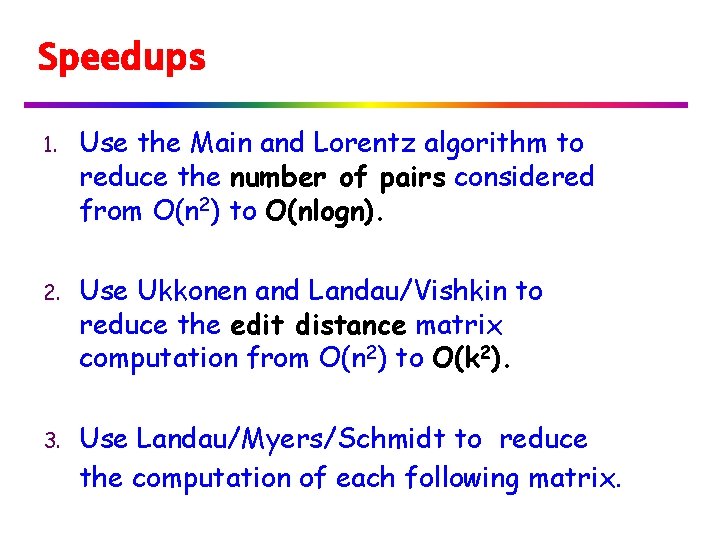
Speedups 1. Use the Main and Lorentz algorithm to reduce the number of pairs considered from O(n 2) to O(nlogn). 2. Use Ukkonen and Landau/Vishkin to reduce the edit distance matrix computation from O(n 2) to O(k 2). 3. Use Landau/Myers/Schmidt to reduce the computation of each following matrix.

Speedup #1: Reduce number of iterations v In first iteration, find all repeats that cross the center of the input string. 1 v Recursively n/2 (center) solve each half. Clearly, there are O(log n) levels. n

To find repeats that cross the center Use the center as an anchor, and pair each index (j) with the center 1 n/2 j Forward Extension: match to the right as much as possible Backward Extension: match to the left as much as possible n

Forward and backward extensions In general: we have a repeat from the leftmost point of the back extension until the rightmost point of the forward extension. 1 n/2 j n

O(n log n) pairs v At each iteration, every j along string pairs with one center. v There are n positions j and log n iterations.

Speedup #2: Reduce matrix Ukkonnen and Landau/Vishkin: v Only central 2 k+1 diagonals of the original edit distance matrix are necessary because moving off these diagonals costs >k errors. v The lowest row with value e ≤ k on each diagonal is sufficient.

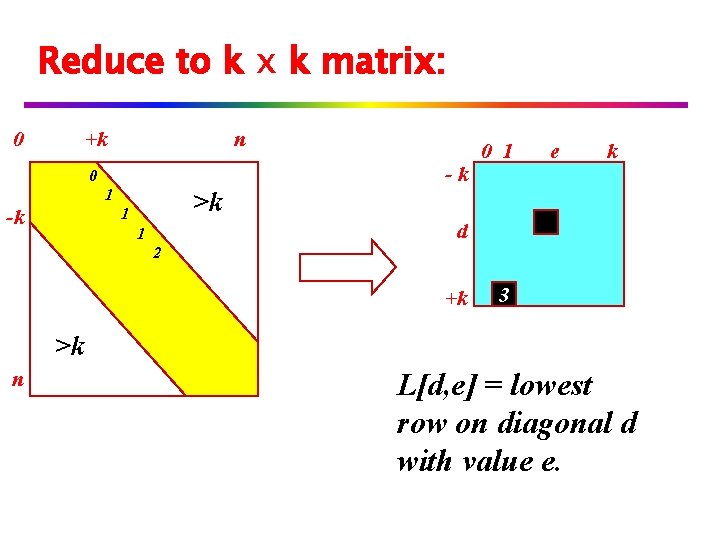
Reduce to k x k matrix: 0 +k n 0 >k 1 1 -k -k 0 1 e k d 1 2 +k 3 >k n L[d, e] = lowest row on diagonal d with value e.

Analysis (so far): v We consider n log n pairs of strings. v But, the edit distance alignment is achieved in nk time per iteration. v Overall: n k log n

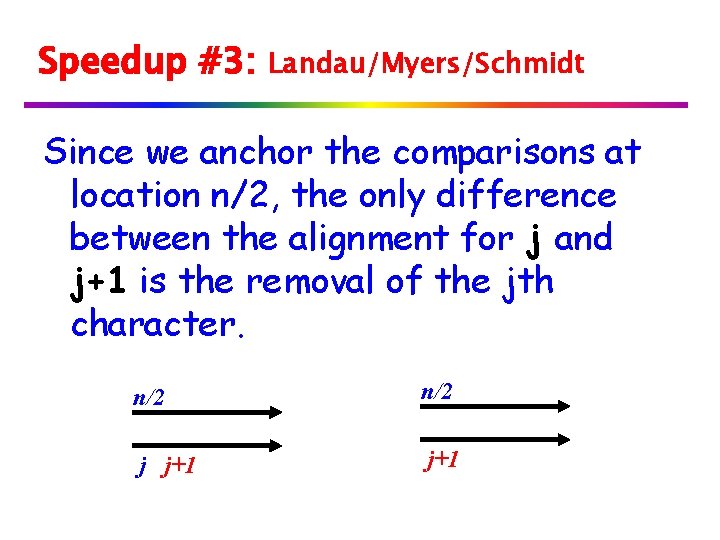
Speedup #3: Landau/Myers/Schmidt Since we anchor the comparisons at location n/2, the only difference between the alignment for j and j+1 is the removal of the jth character. n/2 j j+1

Incremental String Comparison v LMS: in O(k) time we can update the k x k error matrix. v We need an additional O(log k) time to determine the longest possible extension (forward and backward) with a given number of errors.

Time Complexity v Number of iterations: O(log n) v For each iteration: – Compute all first k x k matrices in O(nk) time. – Update for all j in O(k log k) time per j. v Overall: O(nk log n) time. v Space: O(n + k 2).

Software v We implemented this algorithm (without the last step of LMS, and without suffix trees) v The program is efficient, and the repeats found were interesting.

Software

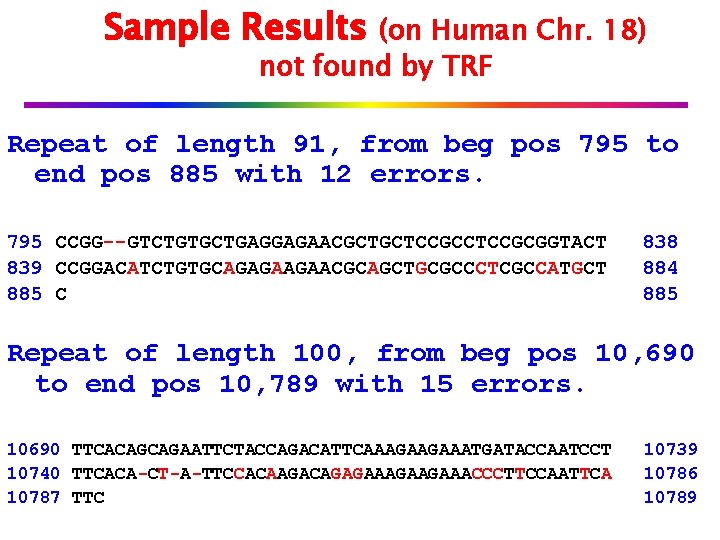
Sample Results (on Human Chr. 18) not found by TRF Repeat of length 91, from beg pos 795 to end pos 885 with 12 errors. 795 CCGG--GTCTGTGCTGAGGAGAACGCTGCTCCGCGGTACT 839 CCGGACATCTGTGCAGAGAAGAACGCAGCTGCGCCCTCGCCATGCT 885 C 838 884 885 Repeat of length 100, from beg pos 10, 690 to end pos 10, 789 with 15 errors. 10690 TTCACAGCAGAATTCTACCAGACATTCAAAGAAGAAATGATACCAATCCT 10740 TTCACA-CT-A-TTCCACAAGACAGAGAAGAAACCCTTCCAATTCA 10787 TTC 10739 10786 10789

Current Work v Since many overlapping repeats are found, we are currently developing criteria for combining and filtering the found repeats.

Open Problems v Use general scoring schemes, such as allowing a cheaper penalty among purines (A, G) and pyrimidines (C, T). v Allow affine gap penalties, where “opening” a gap costs more than extending a gap.