Relational Taxonomy Tree and Bio Data Base by

Relational Taxonomy Tree and Bio. Data. Base by Huhn-Kie Lee 9/9/2020 1

Part I. Relational Taxonomy Tree 9/9/2020 2

Relational Taxonomy Tree (RTT) • Taxonomic hierachy – Kingdom, phylum, class, order, family, genus, species • Lower level inherits higher level’s property: – Properties may be stored “redundantly” • Siblings differ by some properties: – Properties are “disparate, ” so we need different relation schemes 9/9/2020 3
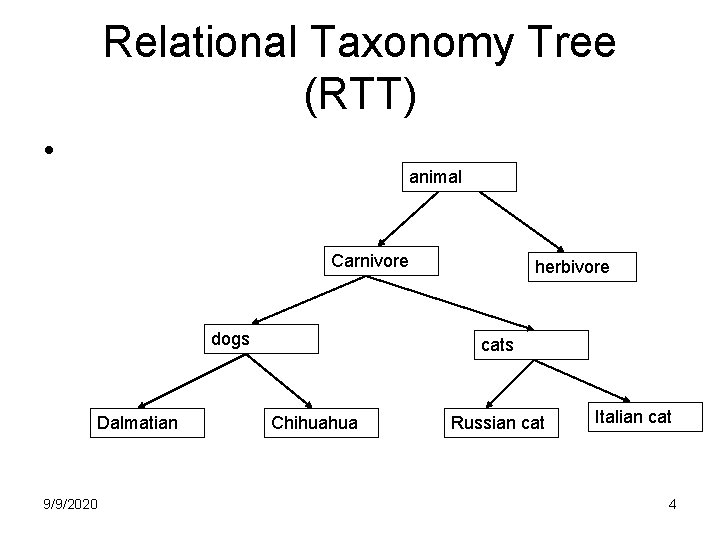
Relational Taxonomy Tree (RTT) • animal Carnivore dogs Dalmatian 9/9/2020 herbivore cats Chihuahua Russian cat Italian cat 4

Relational Taxonomy Tree (RTT) • animal Carnivore (prey, hunting method) dogs Dalmatian 9/9/2020 Herbivore (feeding plant, chewing method) cats Chihuahua Russian cat Italian cat 5
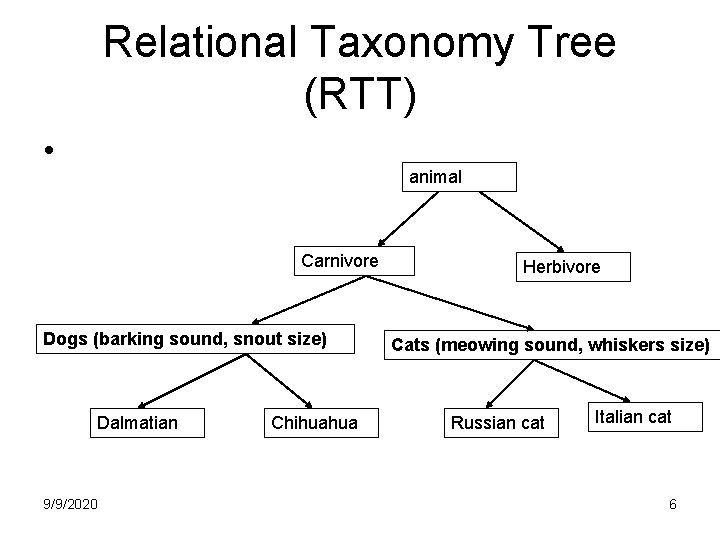
Relational Taxonomy Tree (RTT) • animal Carnivore Dogs (barking sound, snout size) Dalmatian 9/9/2020 Chihuahua Herbivore Cats (meowing sound, whiskers size) Russian cat Italian cat 6
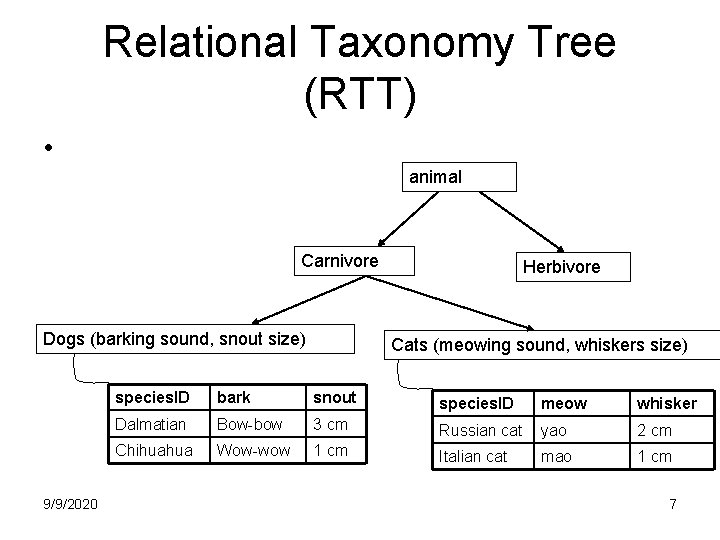
Relational Taxonomy Tree (RTT) • animal Carnivore Dogs (barking sound, snout size) 9/9/2020 Herbivore Cats (meowing sound, whiskers size) species. ID bark snout species. ID meow whisker Dalmatian Bow-bow 3 cm Russian cat yao 2 cm Chihuahua Wow-wow 1 cm Italian cat mao 1 cm 7
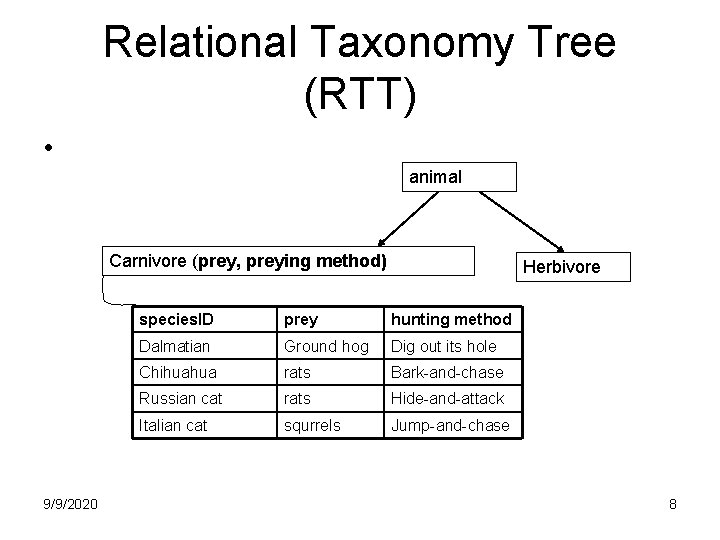
Relational Taxonomy Tree (RTT) • animal Carnivore (prey, preying method) 9/9/2020 Herbivore species. ID prey hunting method Dalmatian Ground hog Dig out its hole Chihuahua rats Bark-and-chase Russian cat rats Hide-and-attack Italian cat squrrels Jump-and-chase 8

Relational Taxonomy Tree (RTT) • Vertical query: – join a relation with its ancestor relation – “Find hunting method of a dog which barks “bow-bow” “ (See relations in slide 6, 5) • SELECT Carn. hunting_method FROM Dogs D, Carnivore Carn WHERE D. species. ID = Carn. species. ID AND D. barking_sound = “bow-bow” 9/9/2020 9

Relational Taxonomy Tree (RTT) • Horizontal query: – join any two relations (may not in same level) – “Find (barking sound, meowing sound) pair of dogs and cats which prey on the same animal (See relations in slide 6, 5) • SELECT D. barking_sound, C. meowing_sound FROM Dogs D, Carnivore Carn 1, Carn 2, Cats C WHERE D. species. ID = Carn 1. species. ID AND C. species. ID = Carn 2. species. ID AND Carn 1. prey = Carn 2. prey 9/9/2020 10
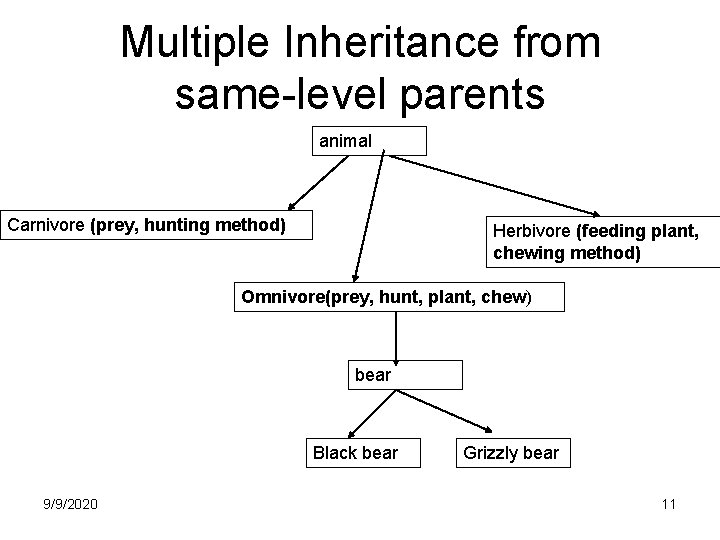
Multiple Inheritance from same-level parents animal Carnivore (prey, hunting method) Herbivore (feeding plant, chewing method) Omnivore(prey, hunt, plant, chew) bear Black bear 9/9/2020 Grizzly bear 11
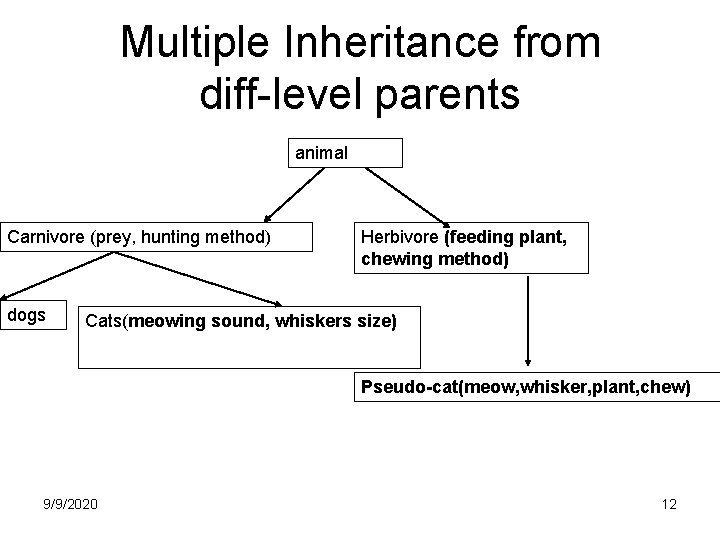
Multiple Inheritance from diff-level parents animal Carnivore (prey, hunting method) dogs Herbivore (feeding plant, chewing method) Cats(meowing sound, whiskers size) Pseudo-cat(meow, whisker, plant, chew) 9/9/2020 12
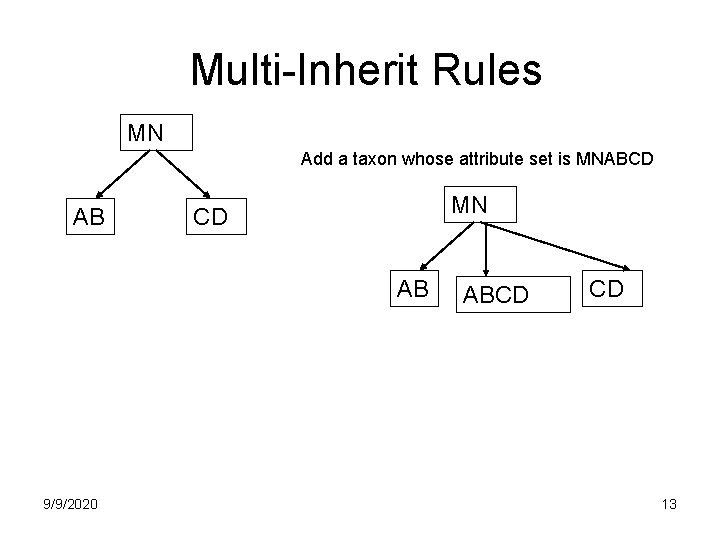
Multi-Inherit Rules MN Add a taxon whose attribute set is MNABCD AB MN CD AB 9/9/2020 ABCD CD 13
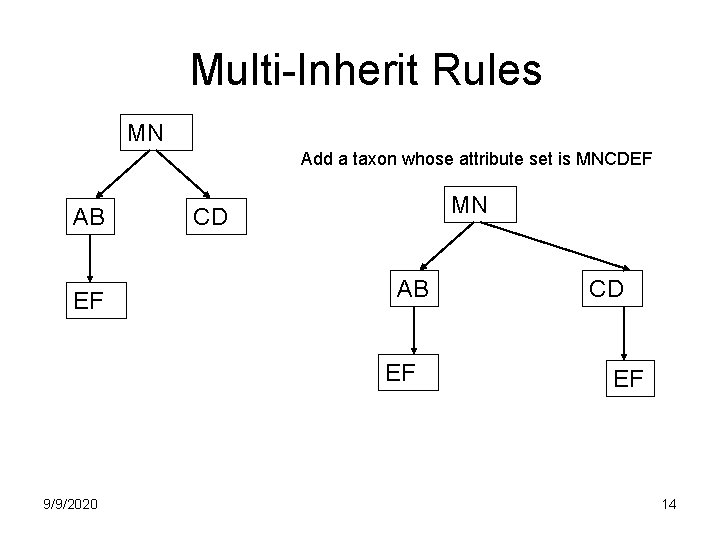
Multi-Inherit Rules MN Add a taxon whose attribute set is MNCDEF AB EF MN CD AB EF 9/9/2020 CD EF 14
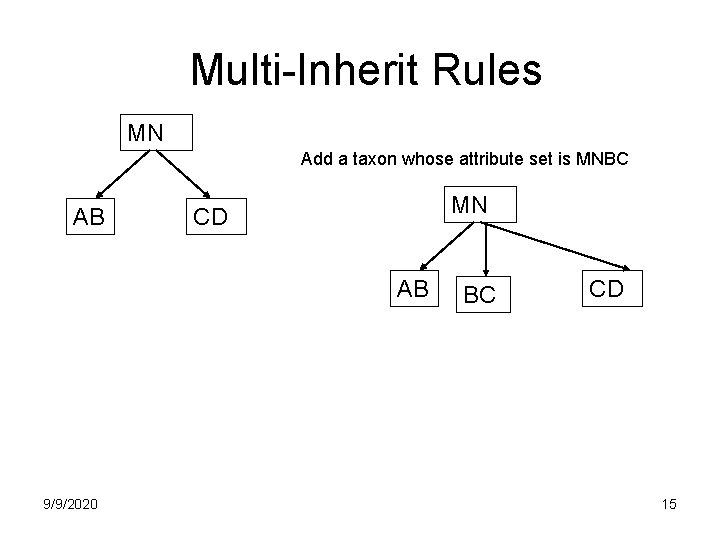
Multi-Inherit Rules MN Add a taxon whose attribute set is MNBC AB MN CD AB 9/9/2020 BC CD 15

Multi-Inherit Rules MN Add a taxon whose attribute set is KL AB CD MN AB 9/9/2020 KL CD 16
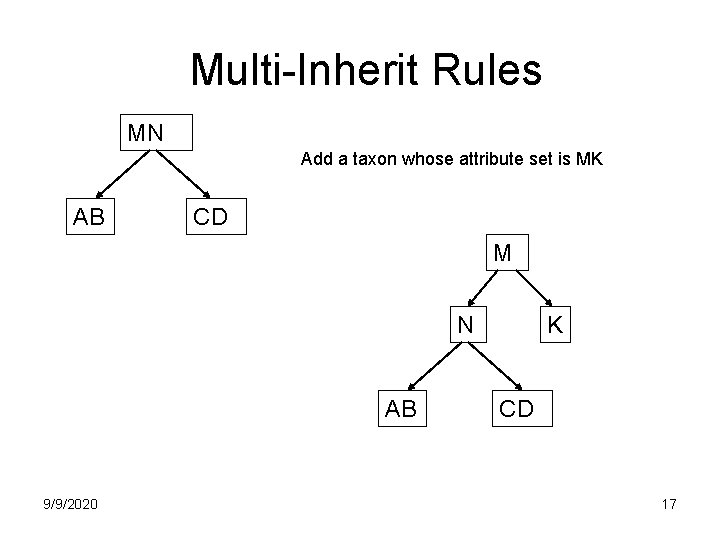
Multi-Inherit Rules MN Add a taxon whose attribute set is MK AB CD M N AB 9/9/2020 K CD 17
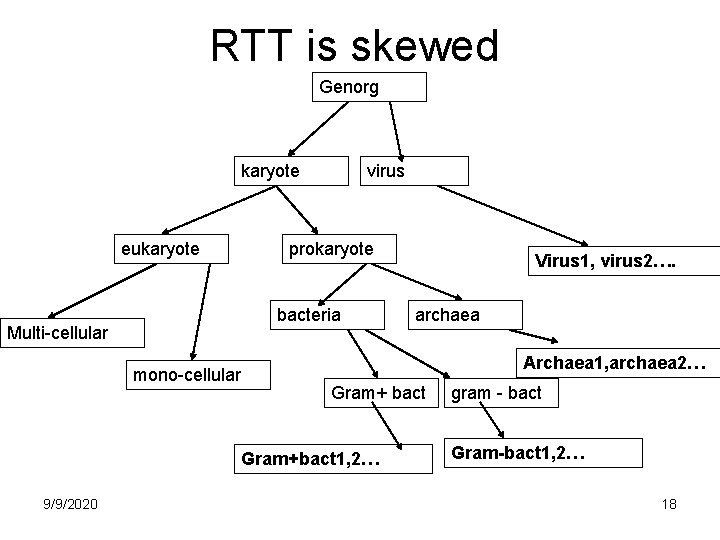
RTT is skewed Genorg karyote eukaryote virus prokaryote bacteria Multi-cellular mono-cellular archaea Archaea 1, archaea 2… Gram+ bact Gram+bact 1, 2… 9/9/2020 Virus 1, virus 2…. gram - bact Gram-bact 1, 2… 18

Terminal Relation Genorg karyote species. ID size AIDS virus 10 nm human 1. 7 m … … virus eukaryote Multi-cellular mono-cellular 9/9/2020 19

Non-terminal Relation Genorg Sub-taxon Ave. size virus 10 nm Karyote 1 m karyote virus eukaryote -Save general trend in Multi-cellular each subtaxon. mono-cellular 9/9/2020 20
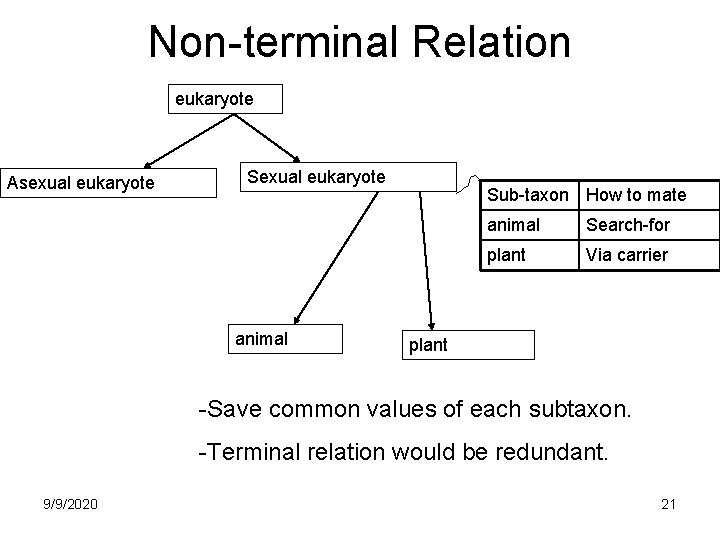
Non-terminal Relation eukaryote Asexual eukaryote Sexual eukaryote animal Sub-taxon How to mate animal Search-for plant Via carrier plant -Save common values of each subtaxon. -Terminal relation would be redundant. 9/9/2020 21

Part II. Bio. Data. Base 9/9/2020 22

Bio. Data. Base (BDB) • Want to store all the information about all the living organisms on the planet – Too many data! – Solution: partition database into “Domains” – Each domain has its own database that stores relevant biological infomation • Want to find correlation between different domains’ information 9/9/2020 23

Bio. Data. Base (BDB) • Consider 3 domains and their relevant info: – Genomics: genes of each species – Ecology: population distribution of species – Environment: a location’s humidity, temperature 9/9/2020 24
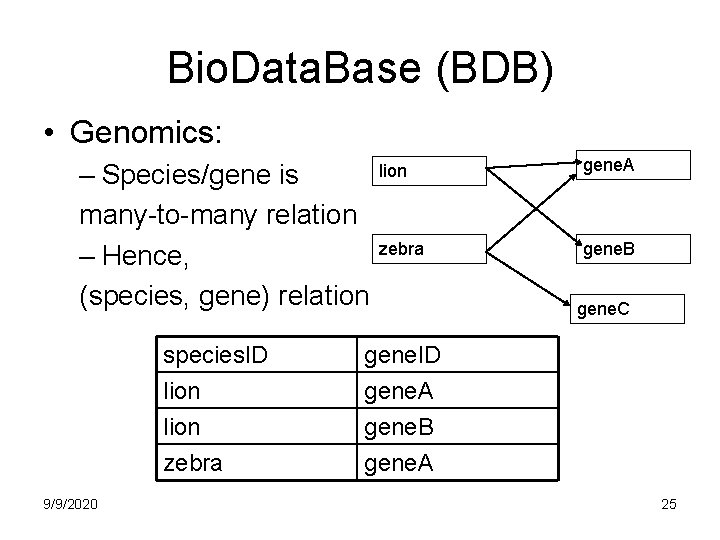
Bio. Data. Base (BDB) • Genomics: – Species/gene is many-to-many relation – Hence, (species, gene) relation species. ID lion zebra 9/9/2020 lion gene. A zebra gene. B gene. C gene. ID gene. A gene. B gene. A 25
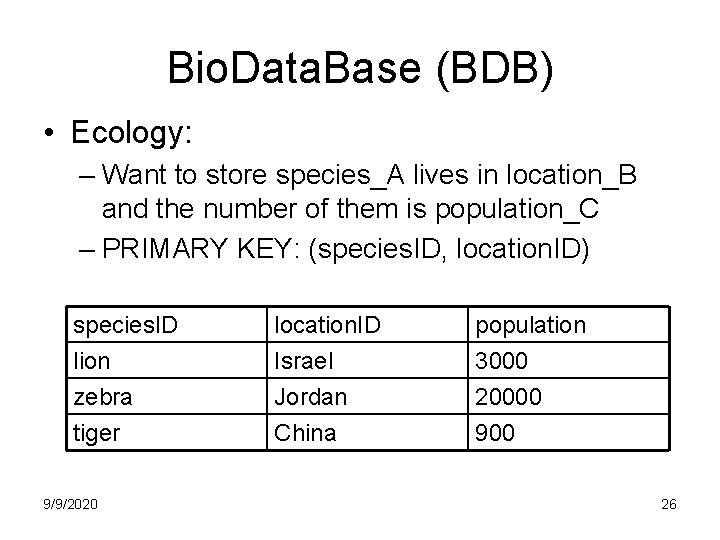
Bio. Data. Base (BDB) • Ecology: – Want to store species_A lives in location_B and the number of them is population_C – PRIMARY KEY: (species. ID, location. ID) species. ID lion zebra tiger 9/9/2020 location. ID Israel Jordan China population 3000 20000 900 26
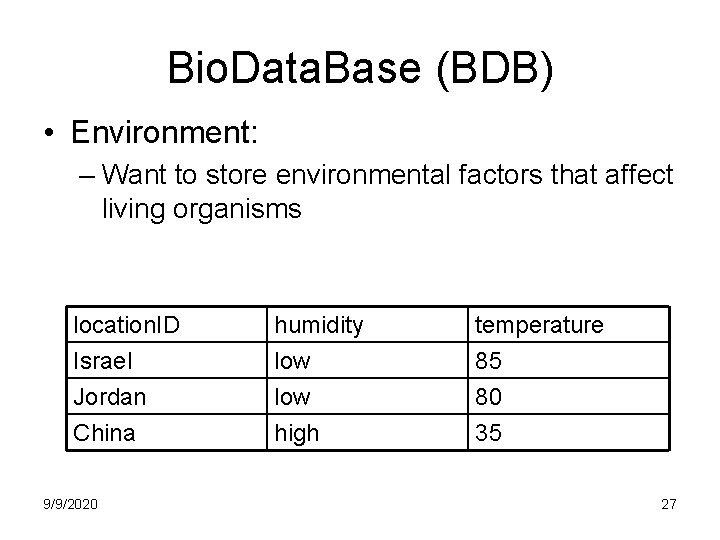
Bio. Data. Base (BDB) • Environment: – Want to store environmental factors that affect living organisms location. ID Israel Jordan China 9/9/2020 humidity low high temperature 85 80 35 27
Bio. Data. Base (BDB) • Want to answer a query that spans all 3 domains: – simply join relations from 3 domains! – “Find genes that are common to (genomics) all species that live in the area (ecology) where humidity is low (environment)” 9/9/2020 28
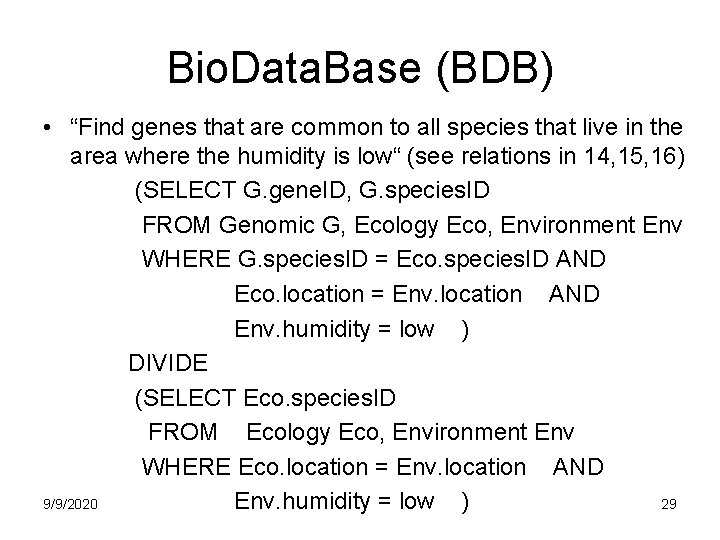
Bio. Data. Base (BDB) • “Find genes that are common to all species that live in the area where the humidity is low“ (see relations in 14, 15, 16) (SELECT G. gene. ID, G. species. ID FROM Genomic G, Ecology Eco, Environment Env WHERE G. species. ID = Eco. species. ID AND Eco. location = Env. location AND Env. humidity = low ) DIVIDE (SELECT Eco. species. ID FROM Ecology Eco, Environment Env WHERE Eco. location = Env. location AND 9/9/2020 29 Env. humidity = low )

Part III. Conclusion & cs 632 Project 9/9/2020 30

Conclusion • Relational Taxonomy Tree solves – Redundancy problem: • diff. species have common attributes. – Disparity problem: • diff. species have diff. attributes • RTT and BDB can serve as the prototype for the infrastructure of the Library of Life Project. 9/9/2020 31

Tentative Project Suggestion • There are four of us: – Helgi, Yoni, Shobhi, mi. • Two of us work on implementation of mini-Relational Taxonomy Tree • The other two of us work implement mini-Bio. Data. Base • All of us implement a program that can process SQL queries on RTT & BDB 9/9/2020 32
So what do you say? 9/9/2020 33
- Slides: 33