Relational Databases Basic Concepts BCHB 524 Lecture 23
Relational Databases: Basic Concepts BCHB 524 Lecture 23 BCHB 524 - Edwards

Outline l l l l What is a (relational) database? When are relational databases used? Commonly used database management systems Using existing databases Creating and populating new databases Python and relational databases Exercises BCHB 524 - Edwards 2

(Relational) Databases l l Databases store information Bioinformatics has lots of file-based information: l l l FASTA sequence databases Genbank format sequences Store sequence, annotation, references, annotation Good as archive or comprehensive reference Poor for a few items Relational databases also store information l l Good for a few items at a time Flexible on which items BCHB 524 - Edwards 3
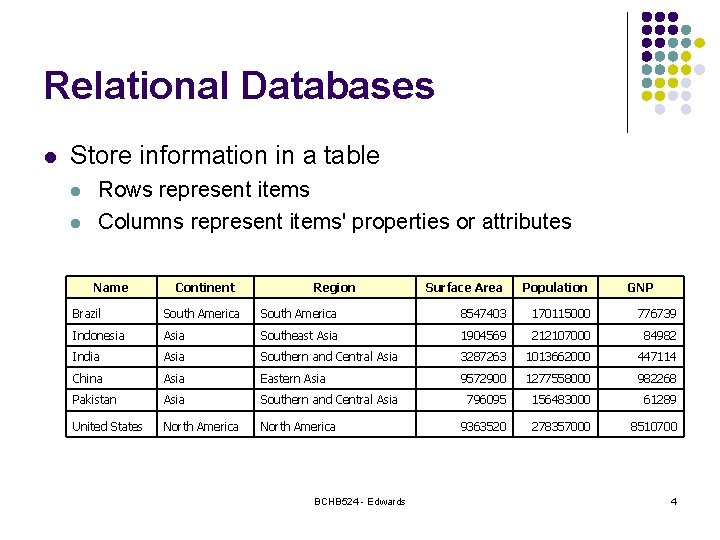
Relational Databases l Store information in a table l l Rows represent items Columns represent items' properties or attributes Name Continent Region Surface Area Population GNP Brazil South America 8547403 170115000 776739 Indonesia Asia Southeast Asia 1904569 212107000 84982 India Asia Southern and Central Asia 3287263 1013662000 447114 China Asia Eastern Asia 9572900 1277558000 982268 Pakistan Asia Southern and Central Asia 796095 156483000 61289 United States North America 9363520 278357000 8510700 BCHB 524 - Edwards 4
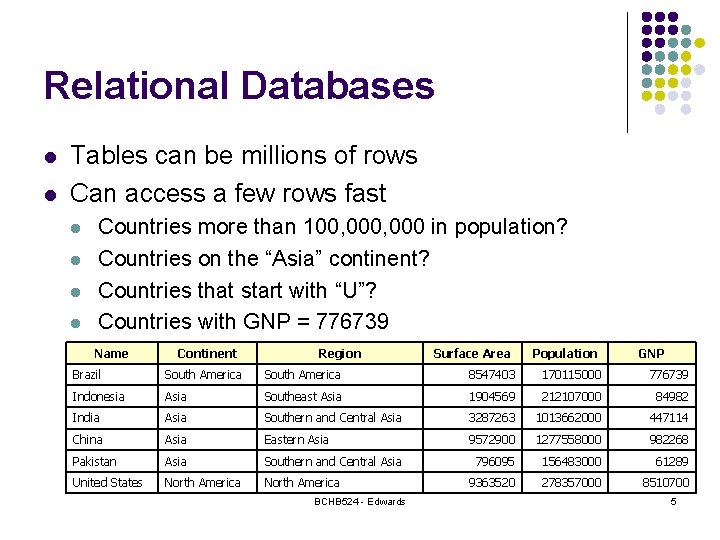
Relational Databases l l Tables can be millions of rows Can access a few rows fast l l Countries more than 100, 000 in population? Countries on the “Asia” continent? Countries that start with “U”? Countries with GNP = 776739 Name Continent Region Surface Area Population GNP Brazil South America 8547403 170115000 776739 Indonesia Asia Southeast Asia 1904569 212107000 84982 India Asia Southern and Central Asia 3287263 1013662000 447114 China Asia Eastern Asia 9572900 1277558000 982268 Pakistan Asia Southern and Central Asia 796095 156483000 61289 United States North America 9363520 278357000 8510700 BCHB 524 - Edwards 5

When are Relational Databases Used? l LARGE datasets l l Store data first. . . l l l For single key, simple data structures often work Store results of expensive compute or data-cleanup l l . . . ask questions later Lookup or sort by many keys l l Does data fit in memory? Compute once and return results many times "Random" or unknown access patterns Specialized data-structures not appropriate l Use string/sequence indexes for sequence data BCHB 524 - Edwards 6
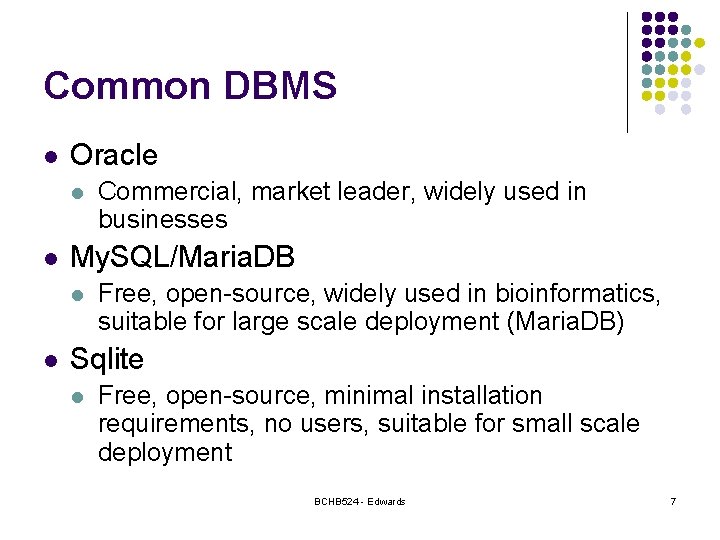
Common DBMS l Oracle l l My. SQL/Maria. DB l l Commercial, market leader, widely used in businesses Free, open-source, widely used in bioinformatics, suitable for large scale deployment (Maria. DB) Sqlite l Free, open-source, minimal installation requirements, no users, suitable for small scale deployment BCHB 524 - Edwards 7

Lets look at some examples l We'll use a third-party program to "look at" Sqlite databases: l l Download examples: l l Sqlite. Studio (Linux), Sqlite. Spy (Windows), … World. db 3, taxa. db 3 from Course data folder Use Sqlite. Studio to look at examples l World. db 3, taxa. db 3 BCHB 524 - Edwards 8

Using existing databases l Use the "select" SQL command to find relevant rows l l l l select * from Country where Population > 10000; select * from Country where Continent = 'Asia'; select * from Country where Name like 'U%'; select * from Country where GNP = 776739; Each command ends in semicolon "; ". "where" specifies the condition/constraint/rule. "*" asks for all attributes from the relevant rows. Lets experiment with world and taxa databases. BCHB 524 - Edwards 9

Using existing databases l Select can combine (“join”) multiple tables l Use the where condition to match rows from each table and “link” corresponding rows… select * from taxonomy, name where taxonomy. rank = 'species' and name_class = 'misspelling' and name. tax_id = taxonomy. tax_id BCHB 524 - Edwards 10

Using existing databases l Select can sort and/or return top 10 select * from taxonomy limit 10; select * from taxonomy order by scientific_name; select * from taxonomy order by tax_id desc limit 10; BCHB 524 - Edwards 11
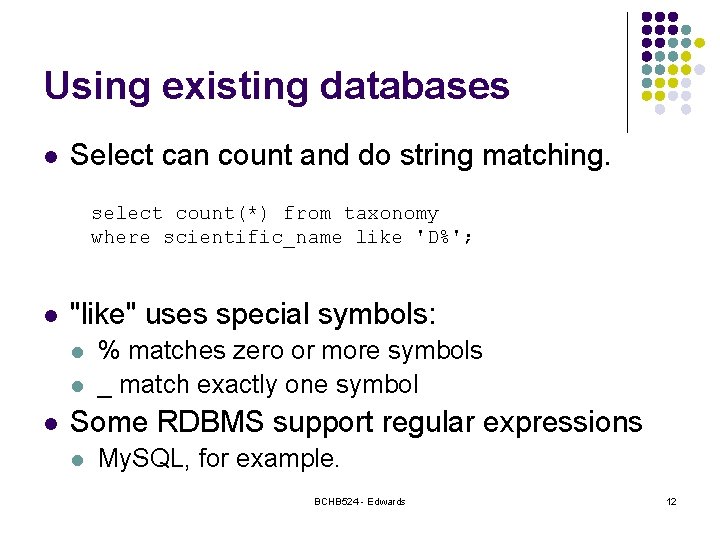
Using existing databases l Select can count and do string matching. select count(*) from taxonomy where scientific_name like 'D%'; l "like" uses special symbols: l l l % matches zero or more symbols _ match exactly one symbol Some RDBMS support regular expressions l My. SQL, for example. BCHB 524 - Edwards 12
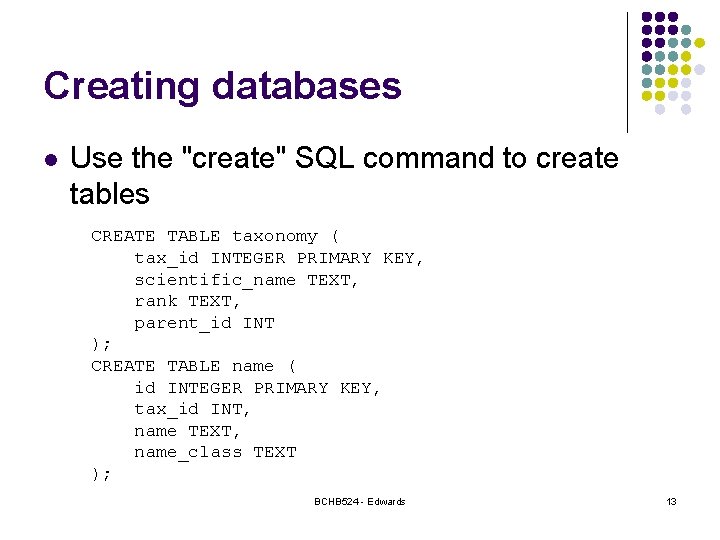
Creating databases l Use the "create" SQL command to create tables CREATE TABLE taxonomy ( tax_id INTEGER PRIMARY KEY, scientific_name TEXT, rank TEXT, parent_id INT ); CREATE TABLE name ( id INTEGER PRIMARY KEY, tax_id INT, name TEXT, name_class TEXT ); BCHB 524 - Edwards 13
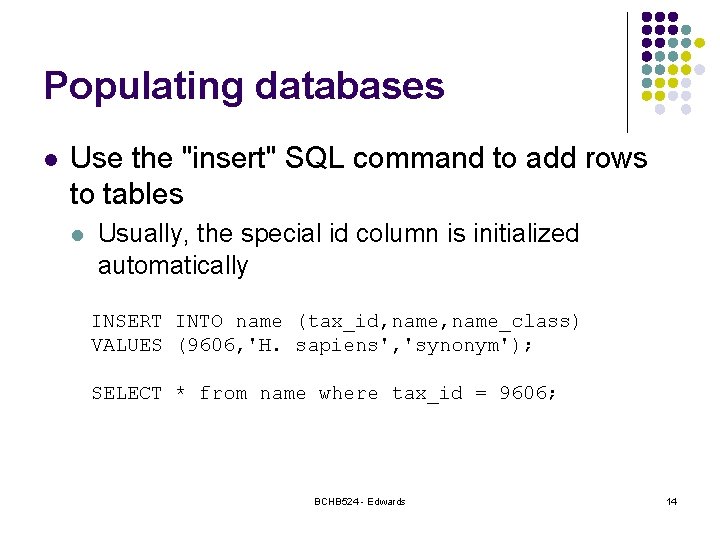
Populating databases l Use the "insert" SQL command to add rows to tables l Usually, the special id column is initialized automatically INSERT INTO name (tax_id, name_class) VALUES (9606, 'H. sapiens', 'synonym'); SELECT * from name where tax_id = 9606; BCHB 524 - Edwards 14

Python and Relational Databases l Issue select statements from python and iterate through the results import sqlite 3 conn = sqlite 3. connect('taxa. db 3') c = conn. cursor() c. execute(""" select * from name where name like 'D%' limit 10; """) for row in c: print(row) l Sometimes it is easiest to make Python do some of the work! BCHB 524 - Edwards 15

Python and Relational Databases l Use parameter substitution for run-time values import sys import sqlite 3 tid = int(sys. argv[1]) conn = sqlite 3. connect('taxa. db 3') params = [tid, 'scientific name'] c = conn. cursor() c. execute(""" select * from name where tax_id = ? and name_class = ? ; """, params) for row in c: BCHB 524 - Edwards print(row) 16

Next-time: Object-relational mappers l Setup python to treat tables as classes, rows as objects # Set up data-model from model import * hs = Taxonomy. get(9606) for n in hs. names: print(n. name, "|", n. name. Class) print() for n in Name. select(Name. q. taxa==10090): print(n. name, "|", n. name. Class) print() condition = Name. q. name. startswith('Da') for n in Name. select(condition): print(n. name, "|", n. name. Class) BCHB 524 - Edwards 17

Exercises l Read through an online course in SQL l l sqlcourse. com, sql-tutorial. net, . . . Write a python program to lookup the scientific name for a user-supplied organism name. BCHB 524 - Edwards 18
- Slides: 18