Regulation of Eukaryotic Gene Expression 2019 Key concepts


- Slides: 34

Regulation of Eukaryotic Gene Expression 2019

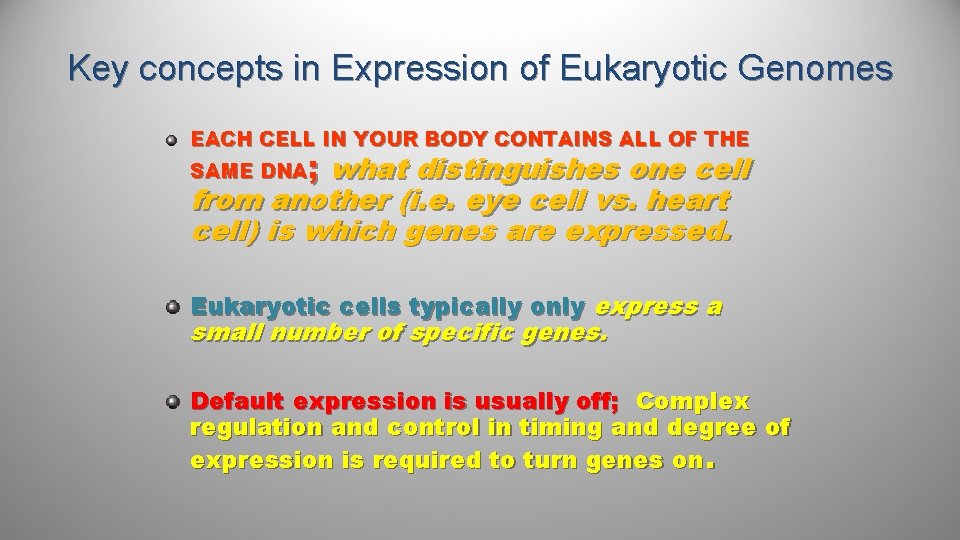
Key concepts in Expression of Eukaryotic Genomes EACH CELL IN YOUR BODY CONTAINS ALL OF THE ; what distinguishes one cell SAME DNA from another (i. e. eye cell vs. heart cell) is which genes are expressed. Eukaryotic cells typically only express a small number of specific genes. Default expression is usually off; Complex regulation and control in timing and degree of expression is required to turn genes on.


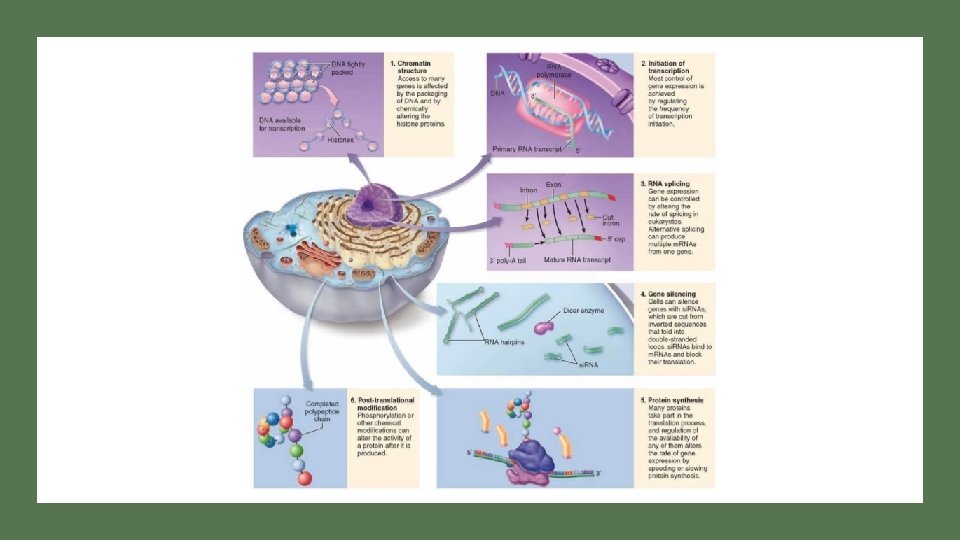
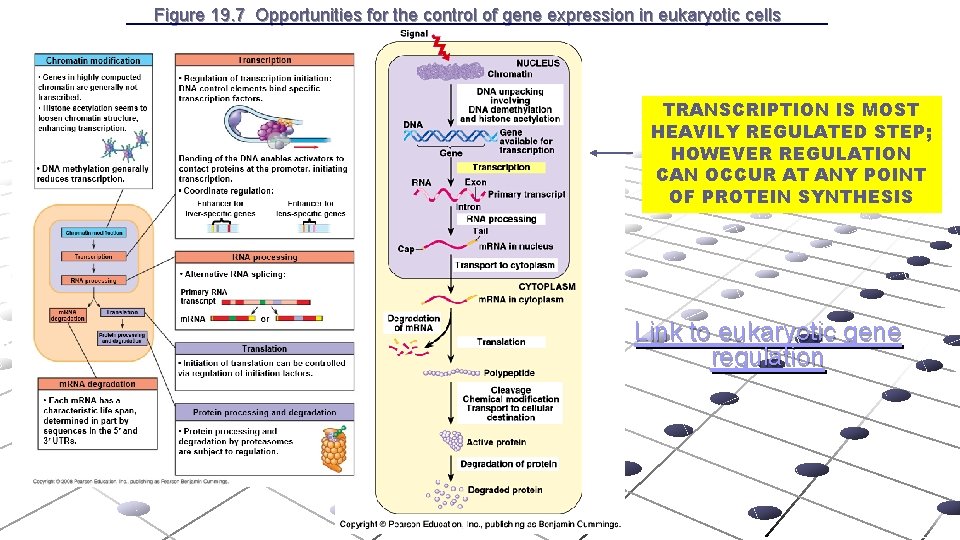
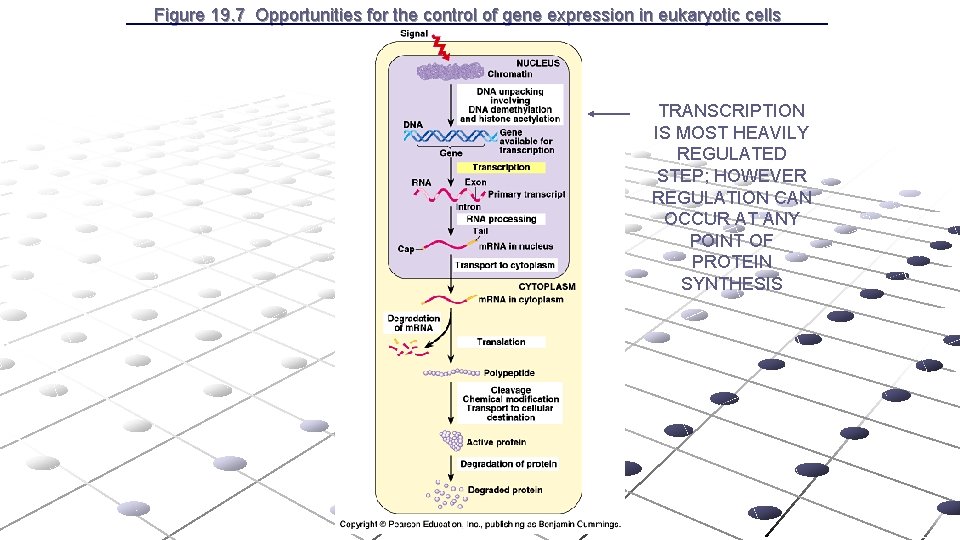
Figure 19. 7 Opportunities for the control of gene expression in eukaryotic cells TRANSCRIPTION IS MOST HEAVILY REGULATED STEP; HOWEVER REGULATION CAN OCCUR AT ANY POINT OF PROTEIN SYNTHESIS Link to eukaryotic gene regulation

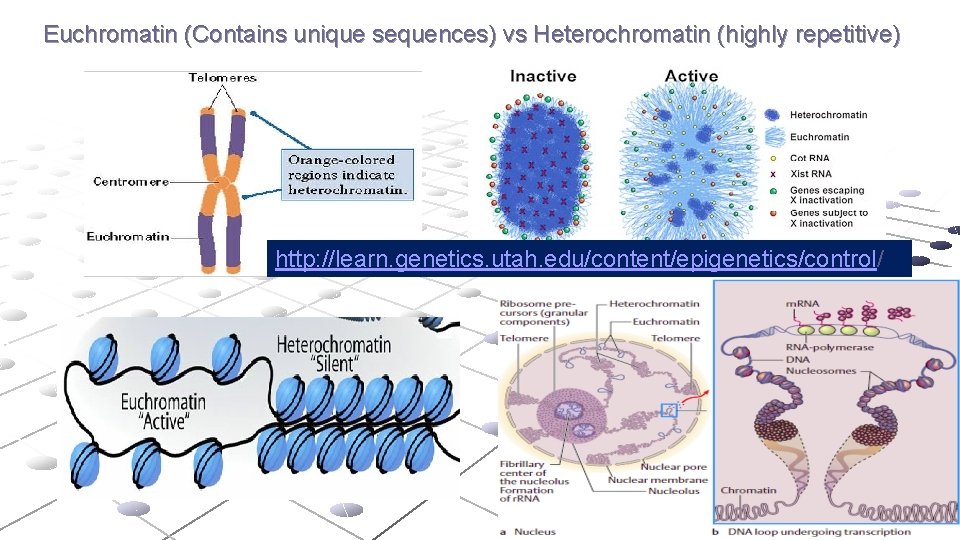
Euchromatin (Contains unique sequences) vs Heterochromatin (highly repetitive) http: //learn. genetics. utah. edu/content/epigenetics/control/

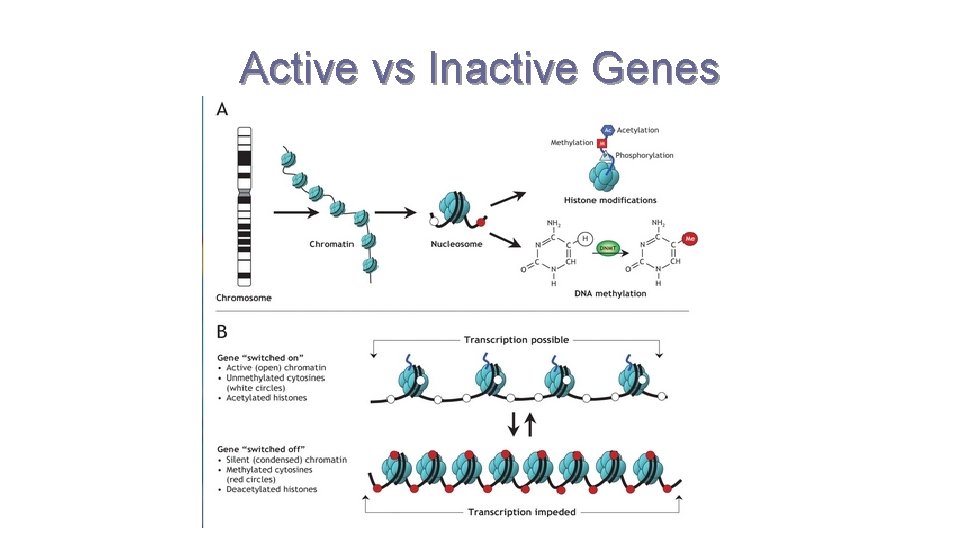
Active vs Inactive Genes

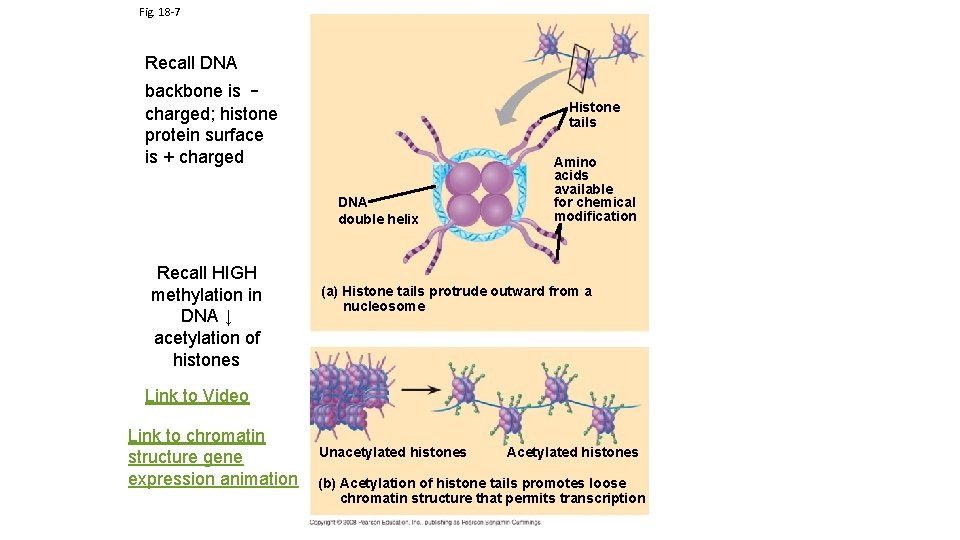
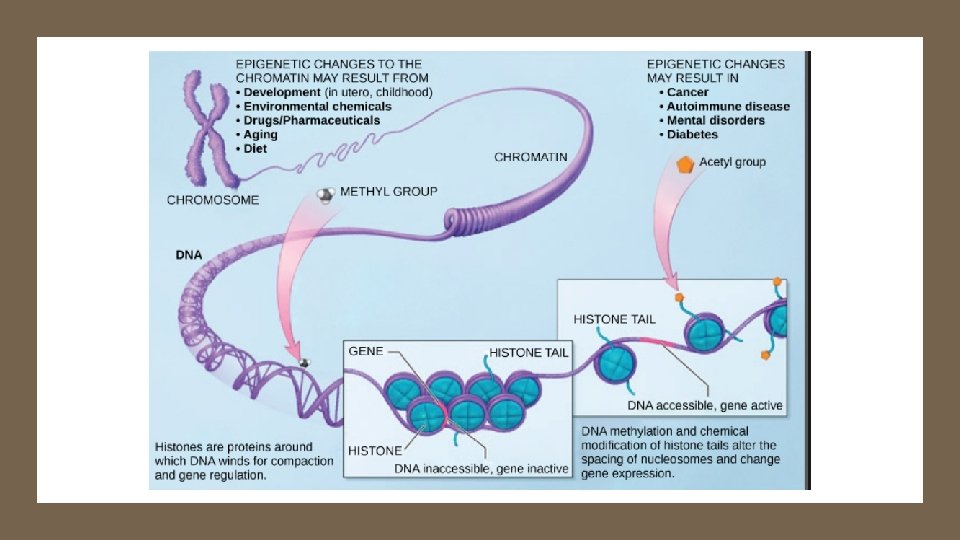
Fig. 18 -7 Recall DNA backbone is charged; histone protein surface is + charged Histone tails DNA double helix Recall HIGH methylation in DNA ↓ acetylation of histones Amino acids available for chemical modification (a) Histone tails protrude outward from a nucleosome Link to Video Link to chromatin structure gene expression animation Unacetylated histones Acetylated histones (b) Acetylation of histone tails promotes loose chromatin structure that permits transcription

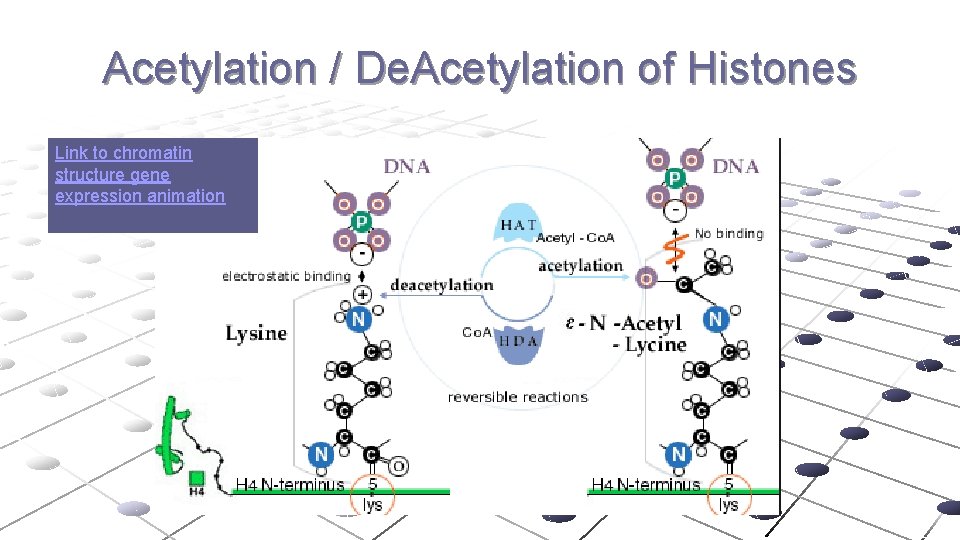
Acetylation / De. Acetylation of Histones Link to chromatin structure gene expression animation

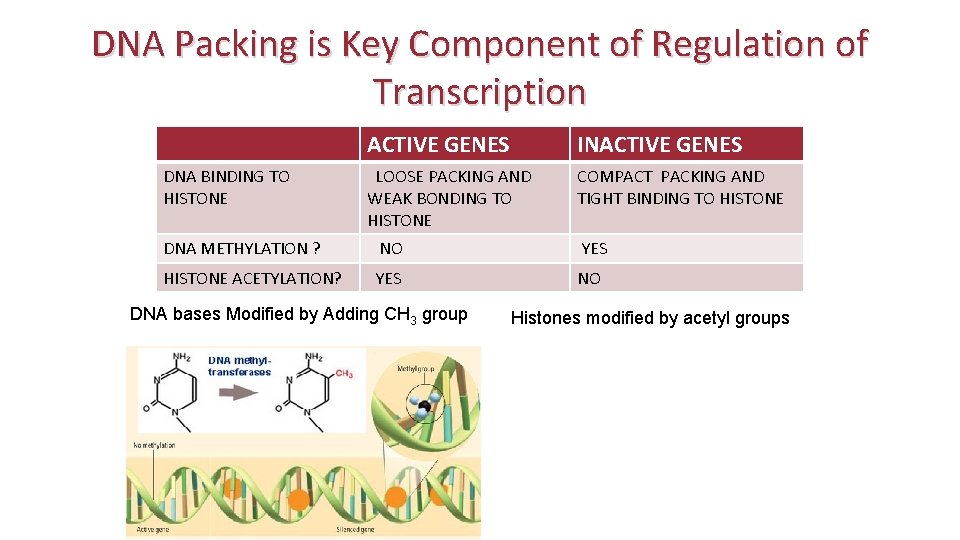
DNA Packing is Key Component of Regulation of Transcription DNA BINDING TO HISTONE ACTIVE GENES INACTIVE GENES LOOSE PACKING AND WEAK BONDING TO HISTONE COMPACT PACKING AND TIGHT BINDING TO HISTONE DNA METHYLATION ? NO YES HISTONE ACETYLATION? YES NO DNA bases Modified by Adding CH 3 group Histones modified by acetyl groups

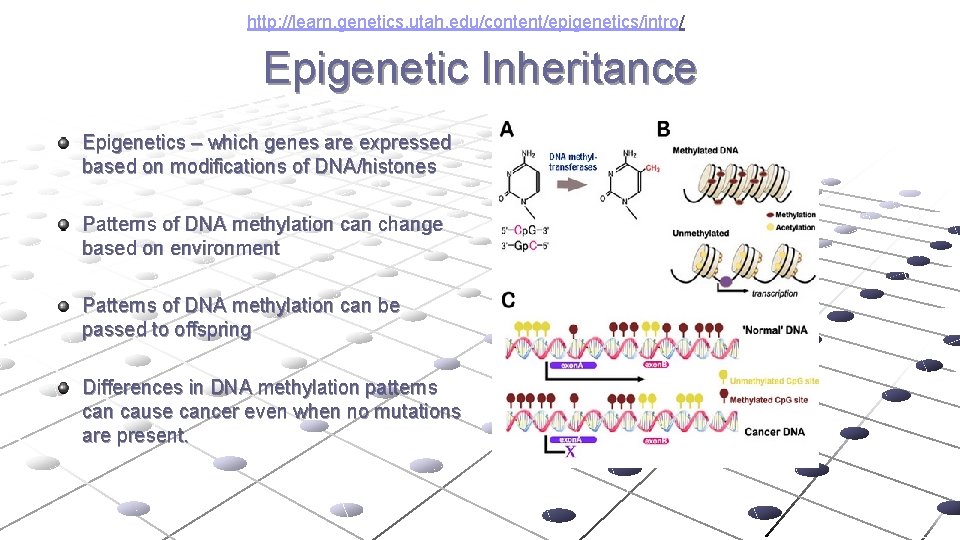
http: //learn. genetics. utah. edu/content/epigenetics/intro/ Epigenetic Inheritance Epigenetics – which genes are expressed based on modifications of DNA/histones Patterns of DNA methylation can change based on environment Patterns of DNA methylation can be passed to offspring Differences in DNA methylation patterns can cause cancer even when no mutations are present.


Review Questions Chromatin What is epigenetics? Ans: Describes which genes are expressed. Describe the how the packing of DNA, level of methylation of bases, and level of histone acetylation impact gene expression. Ø Ans: ACTIVELY TRANSCRIBED GENES - Loosely packed DNA; loose binding between DNA and histone proteins - DNA has LOW level of METHYLATION - Histones have HIGH level of ACETYLATION

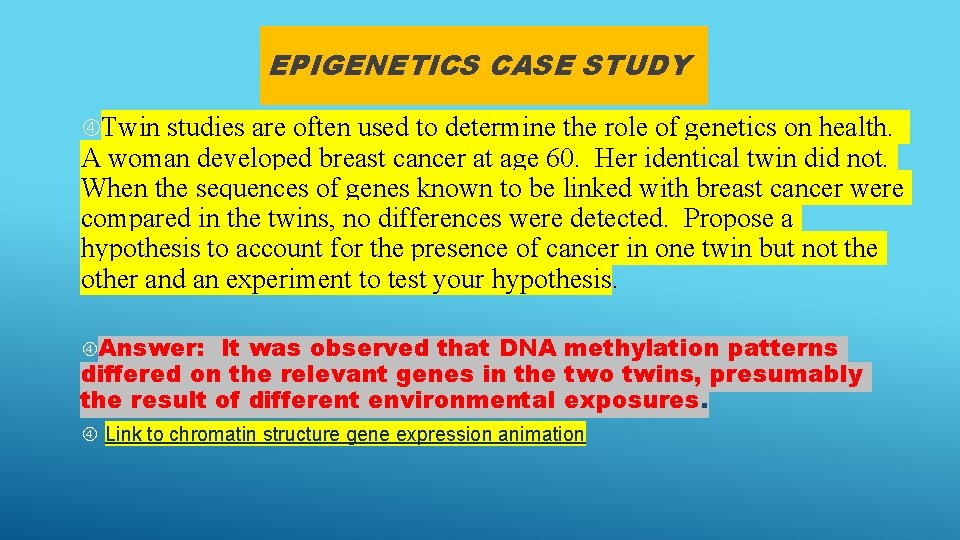
EPIGENETICS CASE STUDY Twin studies are often used to determine the role of genetics on health. A woman developed breast cancer at age 60. Her identical twin did not. When the sequences of genes known to be linked with breast cancer were compared in the twins, no differences were detected. Propose a hypothesis to account for the presence of cancer in one twin but not the other and an experiment to test your hypothesis. Answer: It was observed that DNA methylation patterns differed on the relevant genes in the two twins, presumably the result of different environmental exposures. Link to chromatin structure gene expression animation

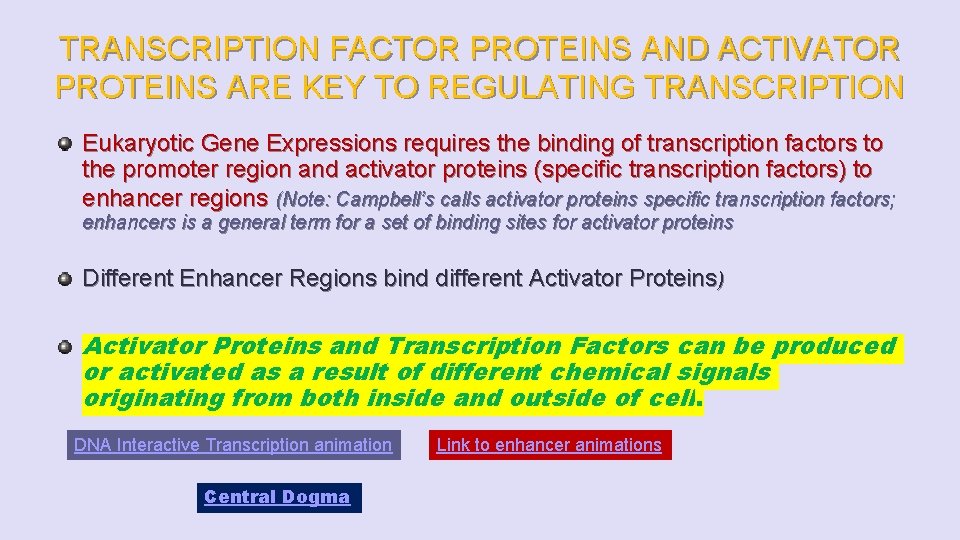
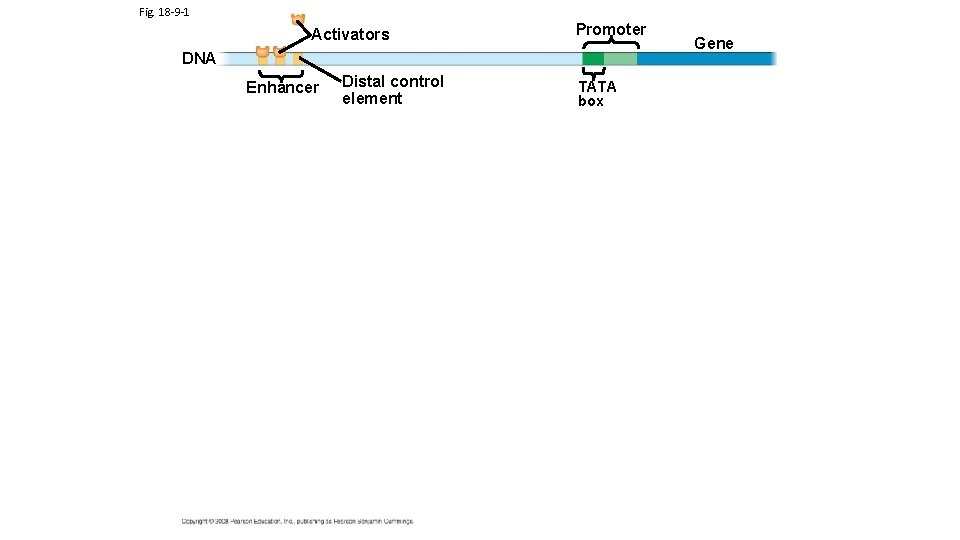
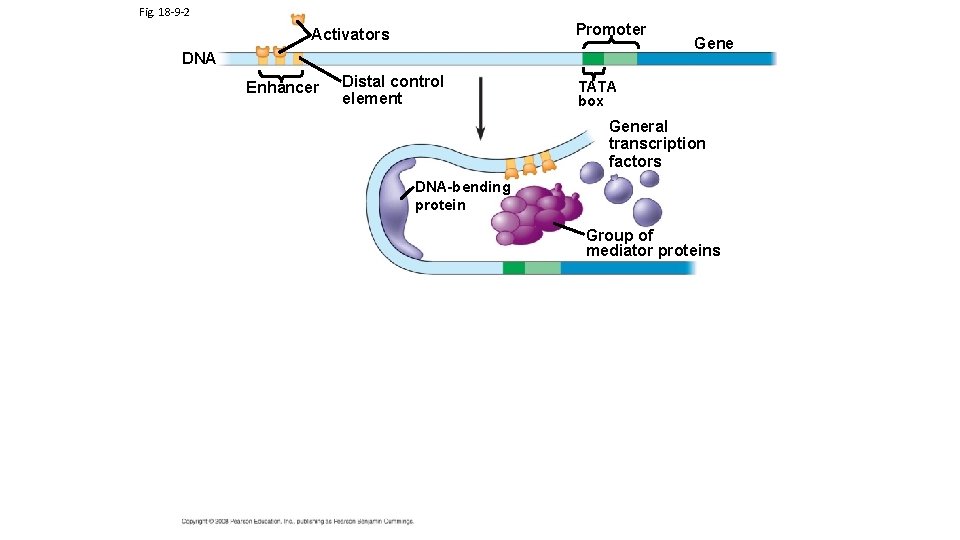
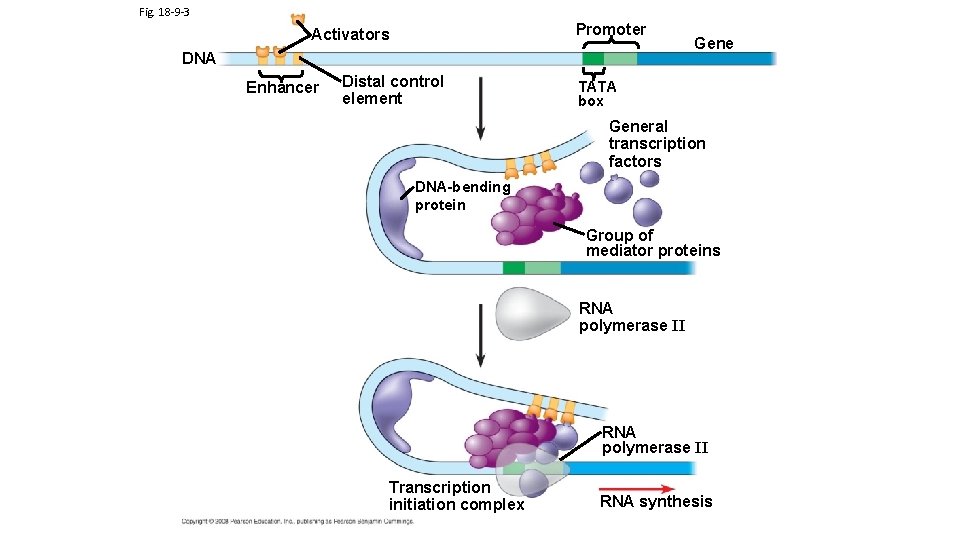
TRANSCRIPTION FACTOR PROTEINS AND ACTIVATOR PROTEINS ARE KEY TO REGULATING TRANSCRIPTION Eukaryotic Gene Expressions requires the binding of transcription factors to the promoter region and activator proteins (specific transcription factors) to enhancer regions (Note: Campbell’s calls activator proteins specific transcription factors; enhancers is a general term for a set of binding sites for activator proteins Different Enhancer Regions bind different Activator Proteins) Activator Proteins and Transcription Factors can be produced or activated as a result of different chemical signals originating from both inside and outside of cell. DNA Interactive Transcription animation Central Dogma Link to enhancer animations

Fig. 18 -9 -1 Activators Promoter DNA Enhancer Distal control element TATA box Gene

Fig. 18 -9 -2 Promoter Activators DNA Enhancer Distal control element Gene TATA box General transcription factors DNA-bending protein Group of mediator proteins

Fig. 18 -9 -3 Promoter Activators DNA Enhancer Distal control element Gene TATA box General transcription factors DNA-bending protein Group of mediator proteins RNA polymerase II Transcription initiation complex RNA synthesis

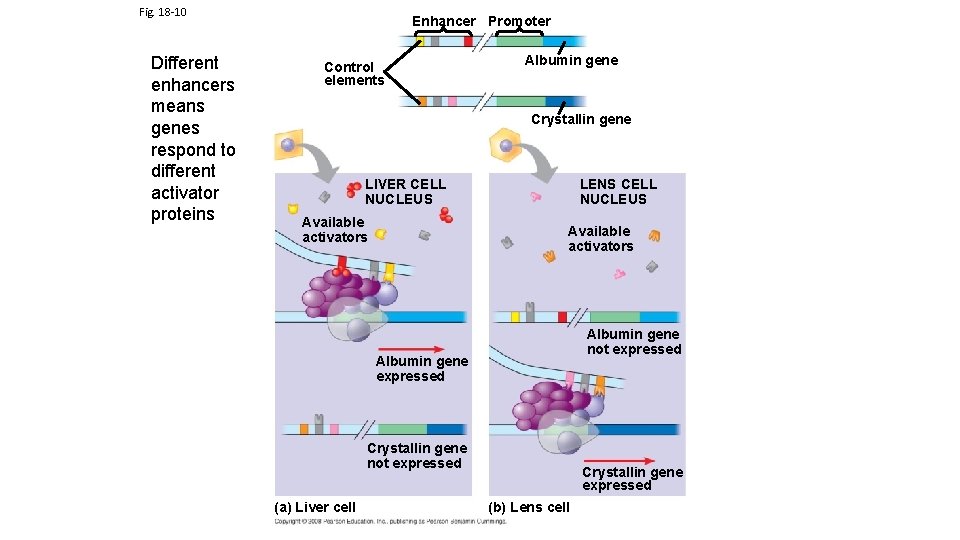
Fig. 18 -10 Different enhancers means genes respond to different activator proteins Enhancer Promoter Control elements Albumin gene Crystallin gene LIVER CELL NUCLEUS Available activators LENS CELL NUCLEUS Available activators Albumin gene not expressed Albumin gene expressed Crystallin gene not expressed (a) Liver cell Crystallin gene expressed (b) Lens cell

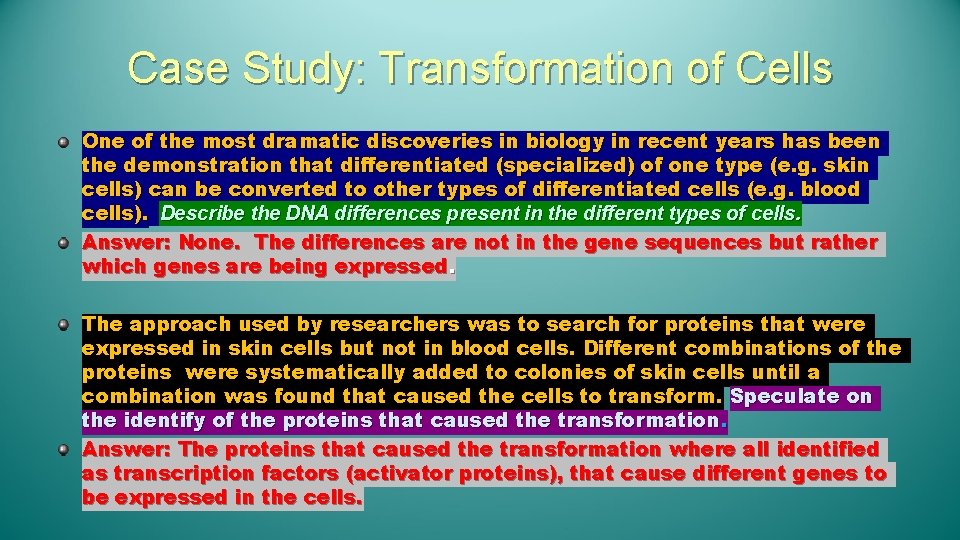
Case Study: Transformation of Cells One of the most dramatic discoveries in biology in recent years has been the demonstration that differentiated (specialized) of one type (e. g. skin cells) can be converted to other types of differentiated cells (e. g. blood cells). Describe the DNA differences present in the different types of cells. Answer: None. The differences are not in the gene sequences but rather which genes are being expressed. The approach used by researchers was to search for proteins that were expressed in skin cells but not in blood cells. Different combinations of the proteins were systematically added to colonies of skin cells until a combination was found that caused the cells to transform. Speculate on the identify of the proteins that caused the transformation. Answer: The proteins that caused the transformation where all identified as transcription factors (activator proteins), that cause different genes to be expressed in the cells.

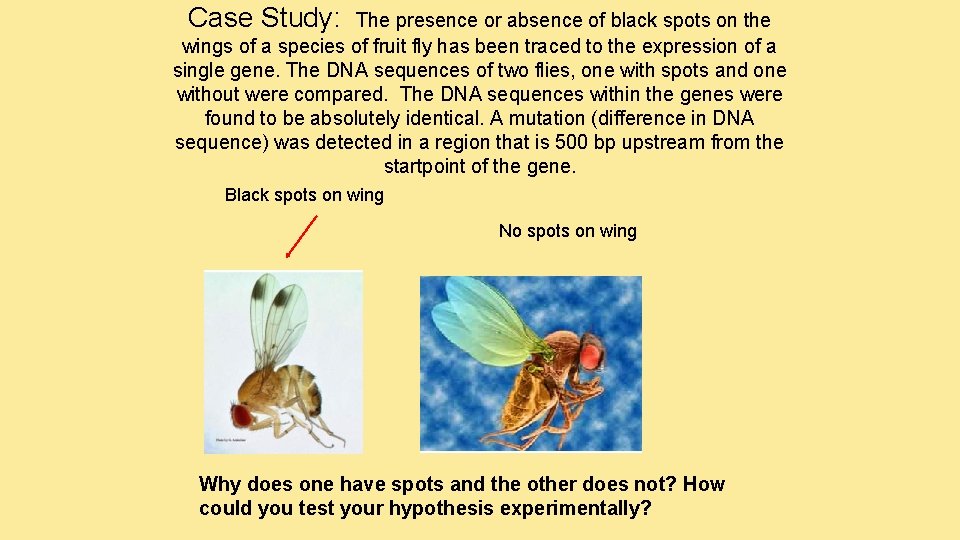
Case Study: The presence or absence of black spots on the wings of a species of fruit fly has been traced to the expression of a single gene. The DNA sequences of two flies, one with spots and one without were compared. The DNA sequences within the genes were found to be absolutely identical. A mutation (difference in DNA sequence) was detected in a region that is 500 bp upstream from the startpoint of the gene. Black spots on wing No spots on wing Why does one have spots and the other does not? How could you test your hypothesis experimentally?

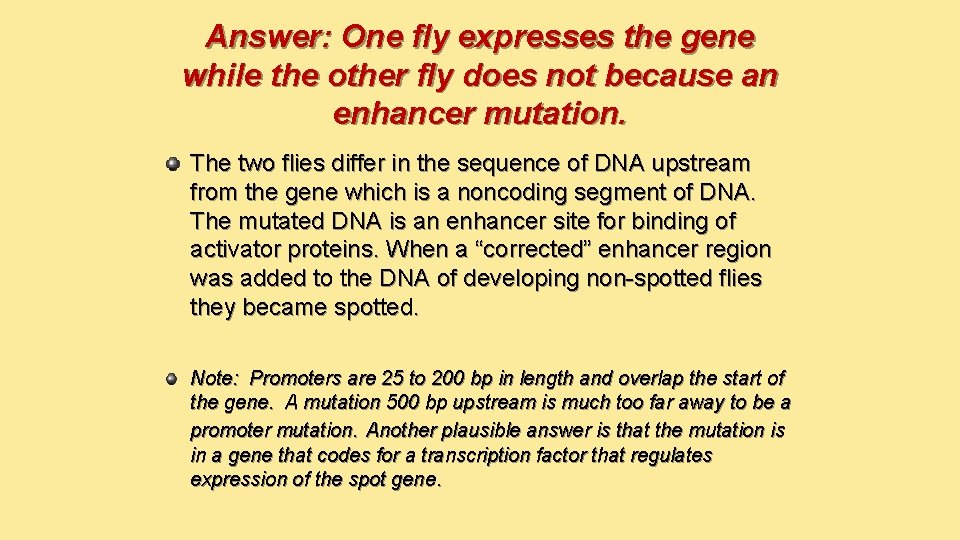
Answer: One fly expresses the gene while the other fly does not because an enhancer mutation. The two flies differ in the sequence of DNA upstream from the gene which is a noncoding segment of DNA. The mutated DNA is an enhancer site for binding of activator proteins. When a “corrected” enhancer region was added to the DNA of developing non-spotted flies they became spotted. Note: Promoters are 25 to 200 bp in length and overlap the start of the gene. A mutation 500 bp upstream is much too far away to be a promoter mutation. Another plausible answer is that the mutation is in a gene that codes for a transcription factor that regulates expression of the spot gene.

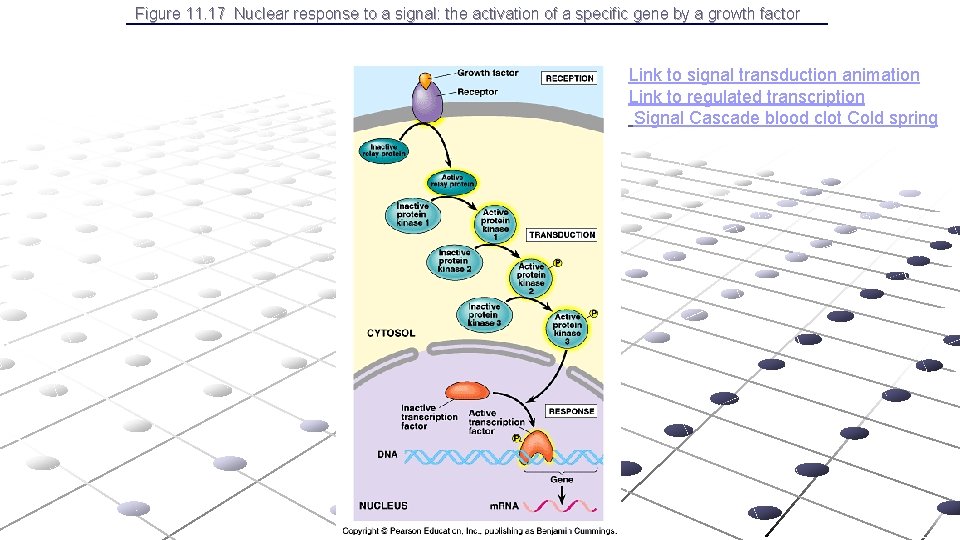
Figure 11. 17 Nuclear response to a signal: the activation of a specific gene by a growth factor Link to signal transduction animation Link to regulated transcription Signal Cascade blood clot Cold spring

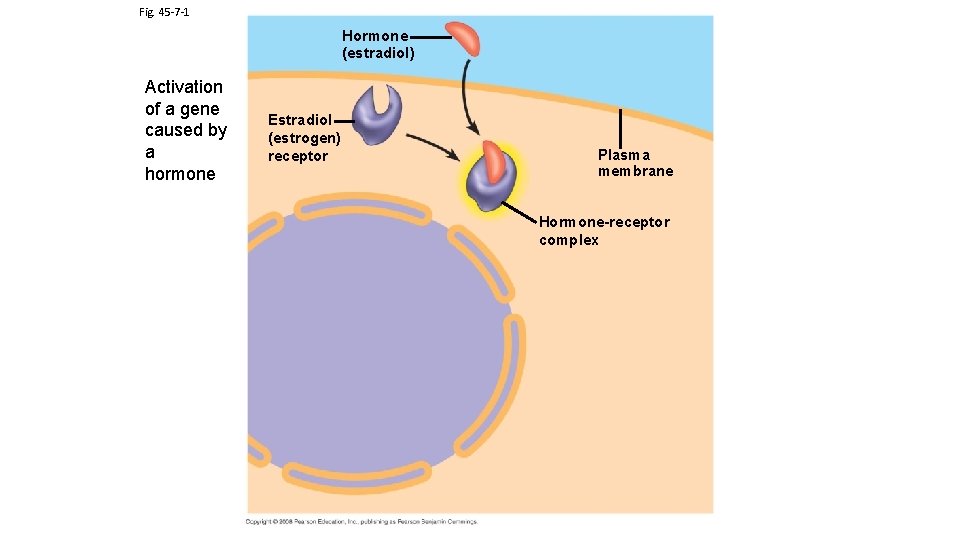
Fig. 45 -7 -1 Hormone (estradiol) Activation of a gene caused by a hormone Estradiol (estrogen) receptor Plasma membrane Hormone-receptor complex

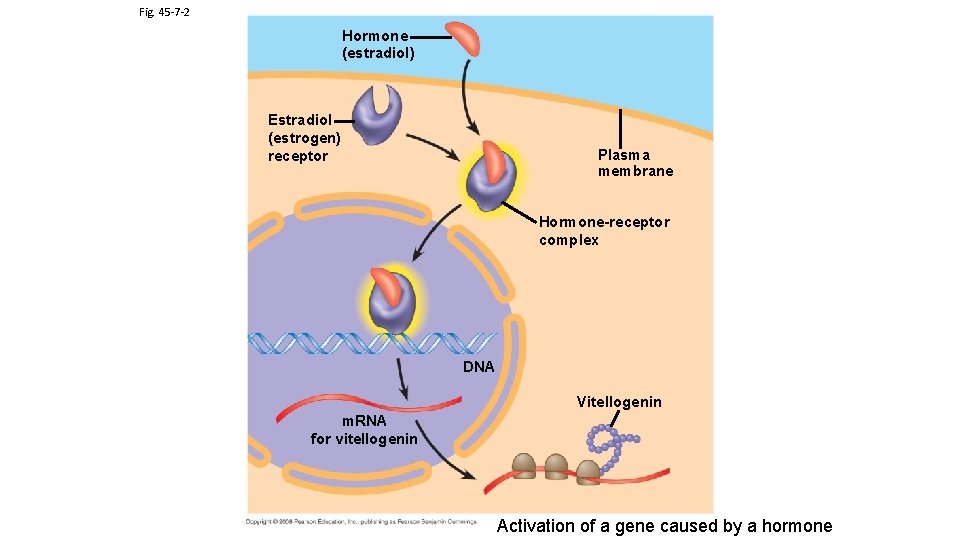
Fig. 45 -7 -2 Hormone (estradiol) Estradiol (estrogen) receptor Plasma membrane Hormone-receptor complex DNA Vitellogenin m. RNA for vitellogenin Activation of a gene caused by a hormone

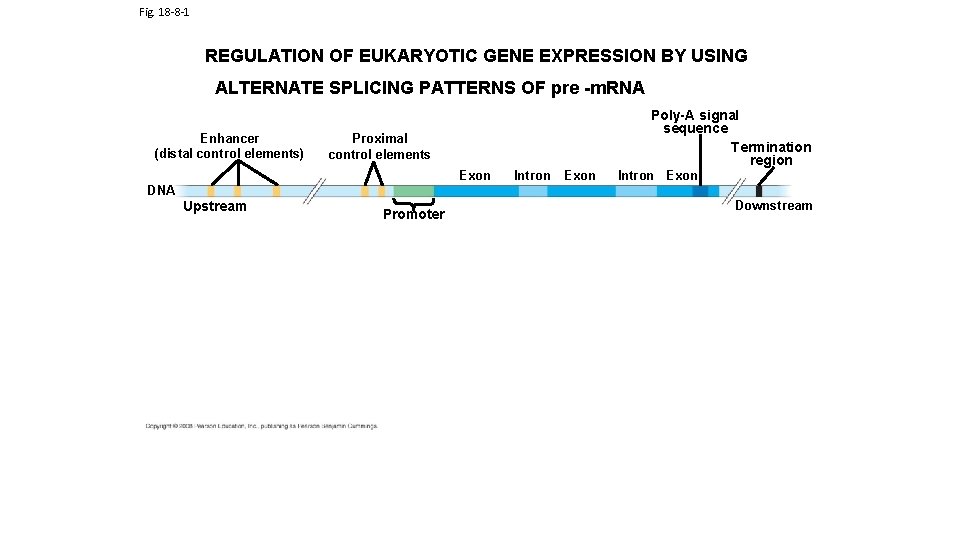
Fig. 18 -8 -1 REGULATION OF EUKARYOTIC GENE EXPRESSION BY USING ALTERNATE SPLICING PATTERNS OF pre -m. RNA Enhancer (distal control elements) Poly-A signal sequence Termination region Proximal control elements Exon DNA Upstream Promoter Intron Exon Downstream

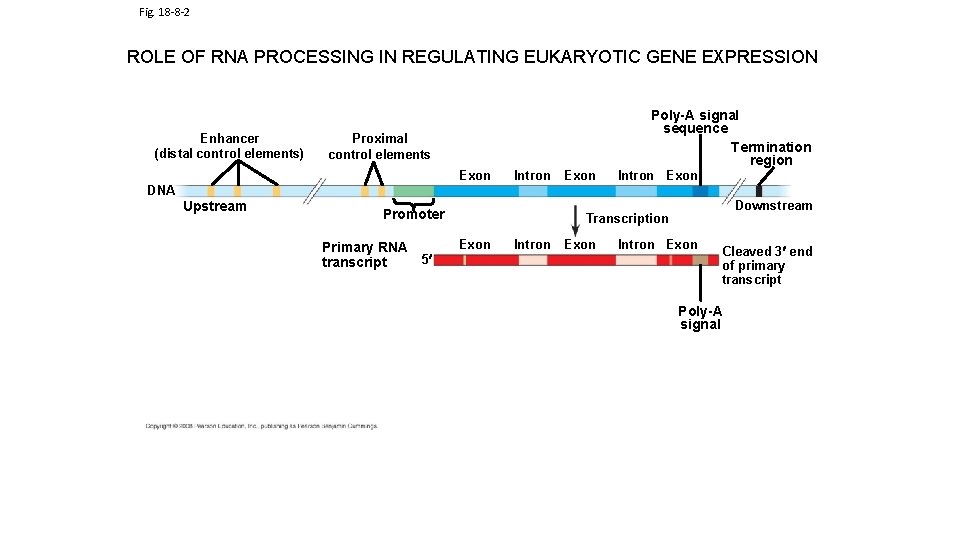
Fig. 18 -8 -2 ROLE OF RNA PROCESSING IN REGULATING EUKARYOTIC GENE EXPRESSION Enhancer (distal control elements) Poly-A signal sequence Termination region Proximal control elements Exon DNA Upstream Intron Promoter Primary RNA 5 transcript Exon Intron Exon Downstream Transcription Exon Intron Exon Cleaved 3 end of primary transcript Poly-A signal

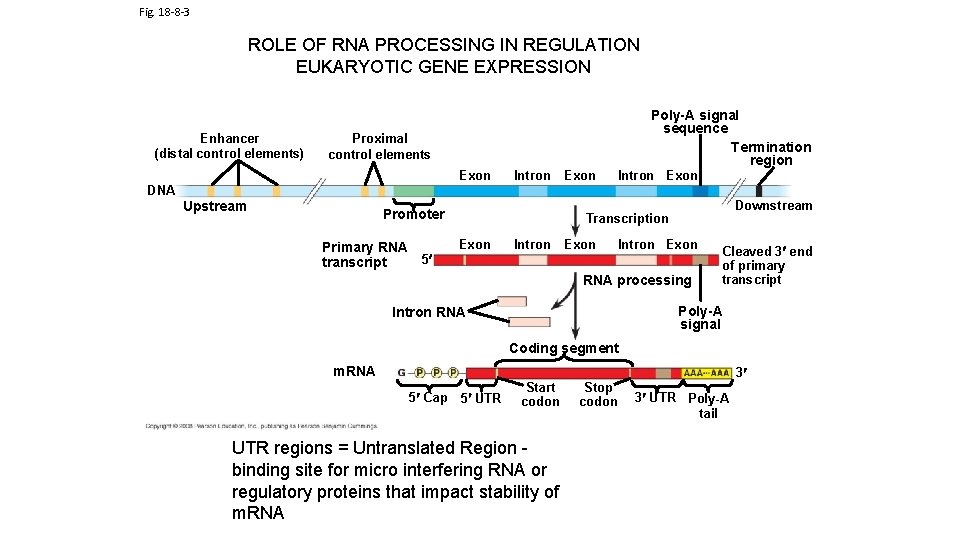
Fig. 18 -8 -3 ROLE OF RNA PROCESSING IN REGULATION EUKARYOTIC GENE EXPRESSION Enhancer (distal control elements) Poly-A signal sequence Termination region Proximal control elements Exon DNA Upstream Intron Promoter Primary RNA 5 transcript Exon Intron Exon Downstream Transcription Exon Intron Exon RNA processing Cleaved 3 end of primary transcript Poly-A signal Intron RNA Coding segment m. RNA 3 5 Cap 5 UTR Start codon UTR regions = Untranslated Region binding site for micro interfering RNA or regulatory proteins that impact stability of m. RNA Stop codon 3 UTR Poly-A tail

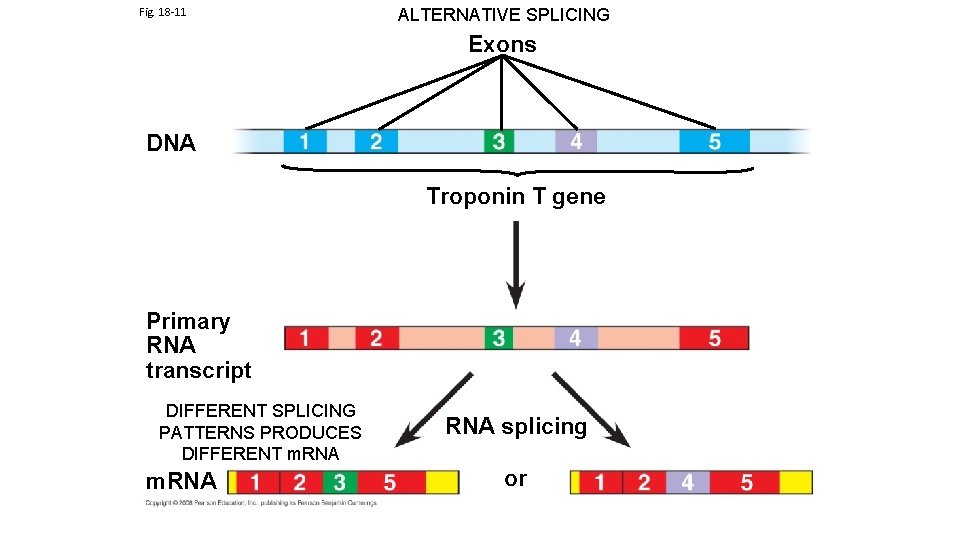
Fig. 18 -11 ALTERNATIVE SPLICING Exons DNA Troponin T gene Primary RNA transcript DIFFERENT SPLICING PATTERNS PRODUCES DIFFERENT m. RNA RNA splicing or

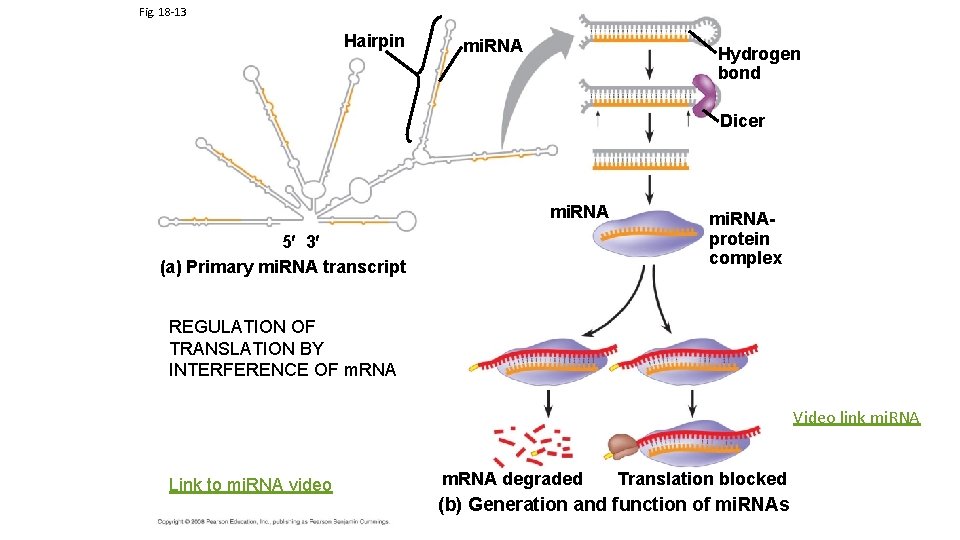
Regulation of Translation Life time of m. RNA is important in determining amount of protein produced. Link to translation regulation animation Rate of Translation is determined in part by the binding of translation initiation protein factors Recent Discoveries suggest that Micro -Interfering RNA (mi. RNA) that is complementary to the m. RNA can also suppress translation

Fig. 18 -13 Hairpin mi. RNA Hydrogen bond Dicer mi. RNA 5 3 (a) Primary mi. RNA transcript mi. RNAprotein complex REGULATION OF TRANSLATION BY INTERFERENCE OF m. RNA Video link mi. RNA Link to mi. RNA video m. RNA degraded Translation blocked (b) Generation and function of mi. RNAs

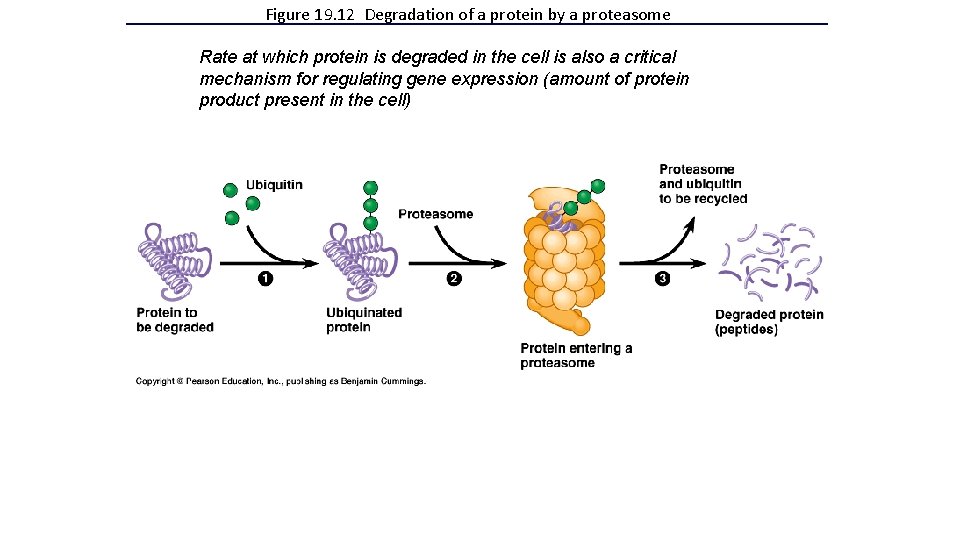
Figure 19. 12 Degradation of a protein by a proteasome Rate at which protein is degraded in the cell is also a critical mechanism for regulating gene expression (amount of protein product present in the cell)

Figure 19. 7 Opportunities for the control of gene expression in eukaryotic cells TRANSCRIPTION IS MOST HEAVILY REGULATED STEP; HOWEVER REGULATION CAN OCCUR AT ANY POINT OF PROTEIN SYNTHESIS

Link to signal transduction animation Link to regulated transcription Signal Cascade blood clot Cold spring

http: //learn. genetics. utah. edu/content/epigenetics/intro/ http: //learn. genetics. utah. edu/content/epigenetics/control/