Recurring conformation of the human immunodeficiency virus type












- Slides: 33

Recurring conformation of the human immunodeficiency virus type 1 gp 120 V 3 loop. Stanfield RL, Ghiara JB, Ollmann Saphire E, Profy AT, and Wilson IA. Virology (2003) Oct 10; 315(1) 159 -73 Bio. Informatics Lab Tuesday, March 9, 2010 Kristoffer Chin Salomon Garcia Michael Piña

Outline • Introduction – Background on HIV Structure – Background on HIV V 3 Region • Materials and Methods – Crystallography • Results and Discussion • References

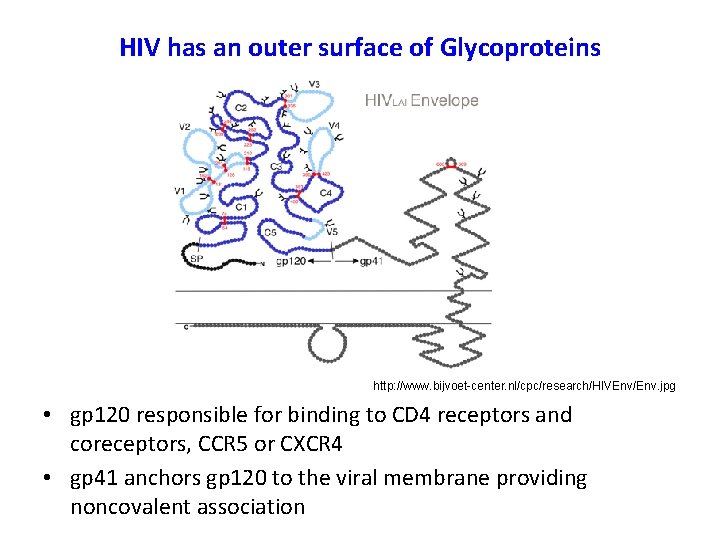
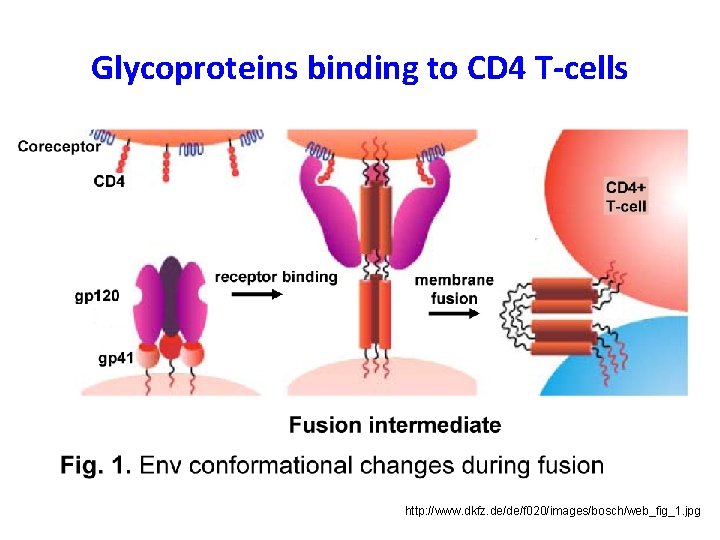
HIV has an outer surface of Glycoproteins http: //www. bijvoet-center. nl/cpc/research/HIVEnv/Env. jpg • gp 120 responsible for binding to CD 4 receptors and coreceptors, CCR 5 or CXCR 4 • gp 41 anchors gp 120 to the viral membrane providing noncovalent association

Glycoproteins binding to CD 4 T-cells http: //www. dkfz. de/de/f 020/images/bosch/web_fig_1. jpg

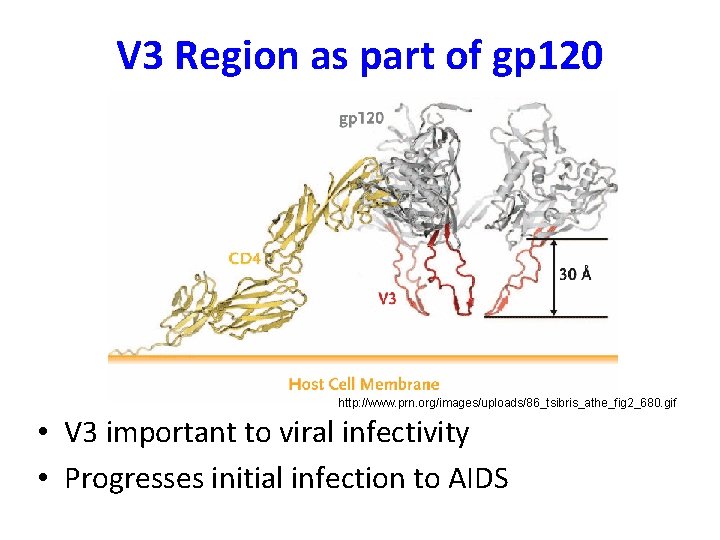
V 3 Region as part of gp 120 http: //www. prn. org/images/uploads/86_tsibris_athe_fig 2_680. gif • V 3 important to viral infectivity • Progresses initial infection to AIDS

Introduction to the Stanfield et al. (2003) Study • V 3 regions have high propensity to elicit neutralizing antibodies • V 3 region inaccessible due to carbohydrates masking or tertiary or quaternary interactions with gp 120 complex • Is there a limited range of conformational states that gp 120 can adopt? • How is V 3 loop recognized by antibodies and how an alteration of sequence, conformation, or exposure can affect it

Finding a dominant conformation for V 3 • Fabs are antibodies that bind to V 3 conformation • Fab 83. 1, 50. 1, and 59. 1 bind to a similar conformation of V 3 region • NMR studies have shown V 3 to form similar hairpin loops • Stabilization of V 3 loops to prevent change of conformation through turns • 5 antibodies used for neutralizing and stablization

Materials and Methods • Mab 83. 1 was made by immunization of an ASW mice with cyclic peptide RP 70 • Antibody was produced in ascites fluid of a mice and purified with an immobilized protein A column • Fab was made from immunoglobin by cleavage • Fab was concentrated to 15. 0 mg/ml for crystallization studies

Fab Purification • Fab was mixed with 16 -mer peptide MP 1 in a 6: 1 mole ration • Crystals were grown using sitting-drop vapor diffusion method with a reservoir solution of 1. 6 M Na/K phosphate, 5% isopropanol, p. H 6. 0. • Crystals grow as clusters of thin plates • Crystals used for this experiments grew over a 2 -week period

Fab Crystallization procedure • Crystals were cryocooled to liquid nitrogen temperatures in order to collect the data in a rapid manner • The crystals were protected by putting them in a solution containing the following 25% glycerol, 1. 6 M Na/K phosphate, 5% isopropanol, p. H 6. 0 • HKL 2000 was used to format all of the data that was obtained

Structure determination • Matthew coefficient was obtained by two Fab molecules • Model was constructed from the constant region of Fab 58. 2 • EPMR program was used to position the model in the cell • EPMR also used to locate the first Fab molecule in the asymetric unit

Stanfield et al. 2003.

Model building and refinement • TOM/FRODO was used to rebuild the mutated hybrid model and to correct the sequence and were subsequently refined with CNS version 1. 1 • Refinement was carried with tight NCS restraints in the beginning and progressively released towards the end of refinement

Structural Analysis • Kabat convention was used to number the molecules – Light and heavy chains are labeled using “L” and “H” – Peptide labeled “P” and was numbered according to HXB 2 isolate sequence • HBPLUS was used to evaluate the Hydrogen bonds • Contacsym program was used to assign van der waals contacts

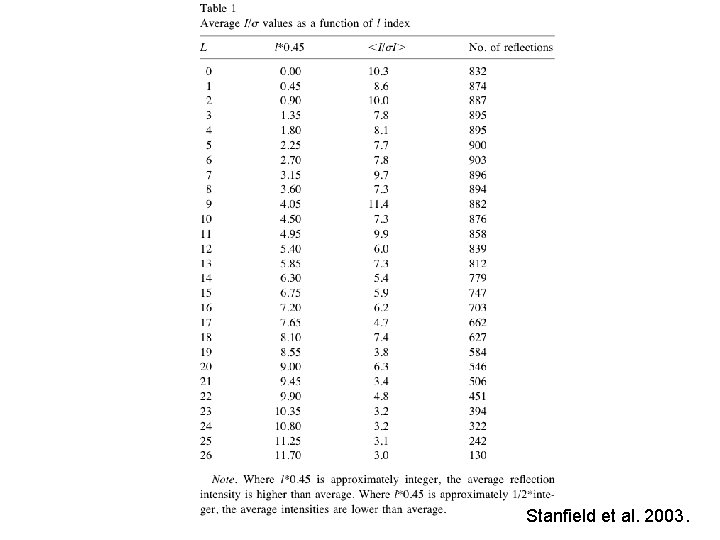
Results and Discussion • Rcryst and Rfree values were slightly higher than other structures determined at 2. 6Å resolution • Electron density maps were good quality • However, repeated refinement and manual rebuilding of the structures caused the higher Rvalues • Rcryst 28. 8%, Rfree 32. 6% • An index (0. 45*l) close to an integer value is strong, whereas close to one-half integer is weak

Stanfield et al. 2003.

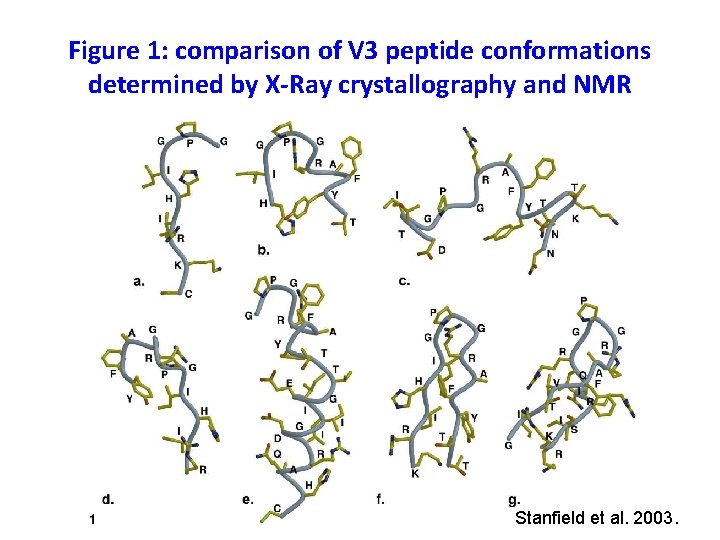
Figure 1: comparison of V 3 peptide conformations determined by X-Ray crystallography and NMR Stanfield et al. 2003.

Figure 2: stereoview of electron density for the V 3 peptide bound to Fab 83. 1 Stanfield et al. 2003.

All CDR loops fall into their expected canonical classes with the exception of L 1 • The L 1 CDR loops have a 5 amino acid insertion after residue L 27 • In both Fabs, the tip of this loop bends away from the antigen binding site in an unusual manner • Comparison with other L 1 loops shows the angle is about 9Å

Figure 3: the structure of the Fab 83. 1 -V 3 peptide complex Stanfield et al. 2003.

Figure 4: Noncanonical loops from Fab 83. 1 Stanfield et al. 2003.

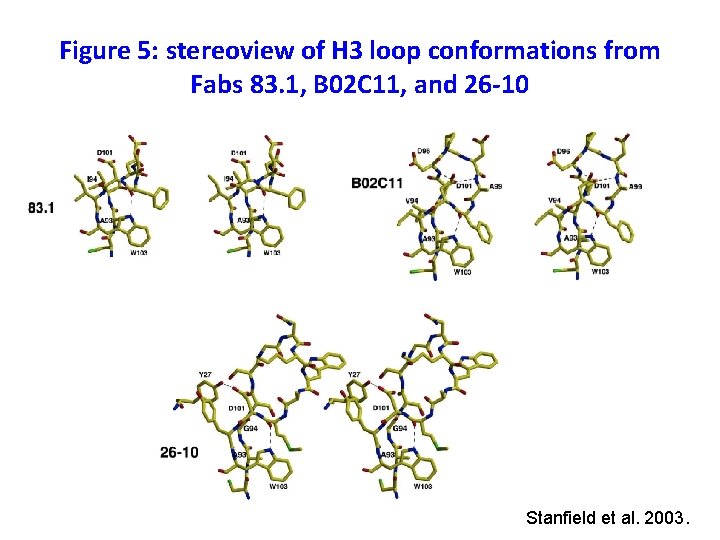
CDR H 3 has a “kinked” base • This was not predicted from its sequence • At least two other Fabs have kinked H 3 bases that were not predicted • Asp. H 101 normally forms a salt bridge (with Arg or Lys), but in this case it does not which is unexpected

Stanfield et al. 2003.

Figure 5: stereoview of H 3 loop conformations from Fabs 83. 1, B 02 C 11, and 26 -10 Stanfield et al. 2003.

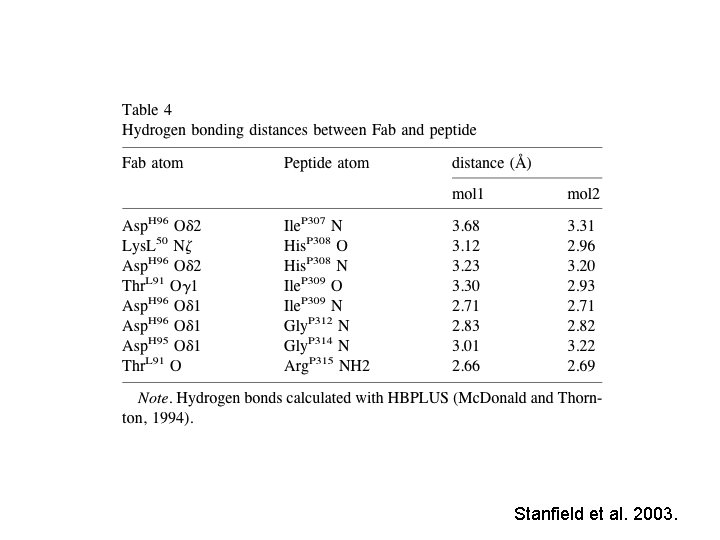
The peptide makes contact with both the light and heavy chains from the Fab • 110 total contacts for 1 molecule • 7 are hydrogen bonds with no charge-charge interactions • 6 hydrogen bonds are to peptide main-chain atoms • 1 bond to Arg side chain • The H 3 CDR makes the most contacts

Stanfield et al. 2003.

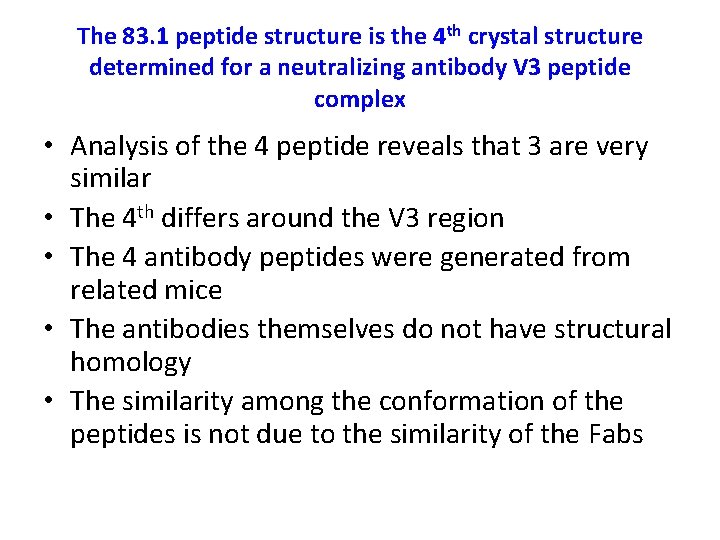
The 83. 1 peptide structure is the 4 th crystal structure determined for a neutralizing antibody V 3 peptide complex • Analysis of the 4 peptide reveals that 3 are very similar • The 4 th differs around the V 3 region • The 4 antibody peptides were generated from related mice • The antibodies themselves do not have structural homology • The similarity among the conformation of the peptides is not due to the similarity of the Fabs

Figure 6: comparison of the V 3 peptide conformations Stanfield et al. 2003.

Figure 7: the antibody combining site of Fab 83. 1 Stanfield et al. 2003.

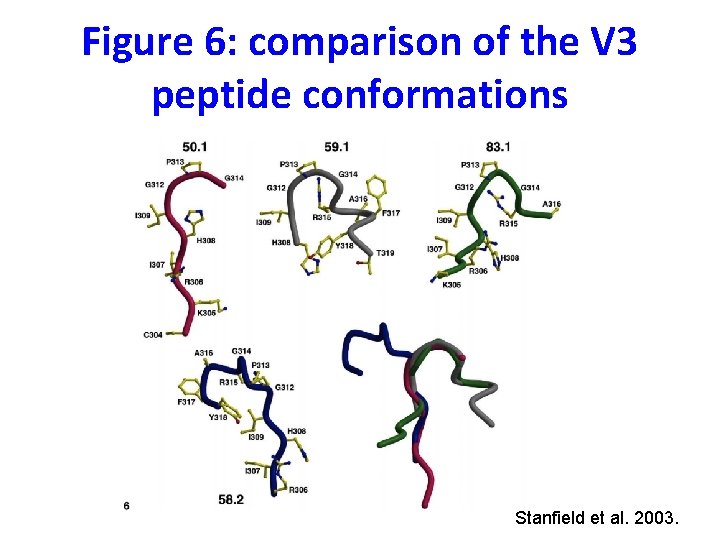
Figure 8: antigen binding sites of Fabs 83. 1, 59. 1, 58. 2, & 50. 1 Stanfield et al. 2003.

The peptides, although adopting the same shapes, bind in different orientations and locations in the antibody • The antibodies were chosen for ability to neutralize (bind to intact viruses) • These peptide conformations should reflect “preferred” conformations of the V 3 loop • The identified V 3 structures represent a recurring conformer on the intact virus

The X-rays of V 3 peptides in complex with antibodies help define the range of V 3 conformation • Studies suggest that V 3 interacts with coreceptors CCR 5 and CXCR 4 during cell entry • This information may be useful in the design of V 3 based inhibitors • Ultimately, a better understanding of the gp 120/gp 41 structure (and the V 3 region) is vital for understanding how HIV-1 carries out its binding and fusion activities

References • Stanfield et al. Recurring conformation of the human immunodeficiency virus type 1 gp 120 V 3 loop. Virology (2003) Oct 10; 315(1) 159 -73 • http: //www. bijvoet-center. nl/cpc/research/HIVEnv/Env. jpg • http: //www. dkfz. de/de/f 020/images/bosch/web_fig_1. jpg • http: //www. prn. org/images/uploads/86_tsibris_athe_fig 2_680. gif
 Secondary immunodeficiency diseases
Secondary immunodeficiency diseases Severe combined immunodeficiency
Severe combined immunodeficiency Bond angle in cyclohexane
Bond angle in cyclohexane Horse conformation pictures
Horse conformation pictures E and z alkene
E and z alkene Tetrahydropyran chair conformation
Tetrahydropyran chair conformation Planar trans-1 2-dimethylcyclohexane
Planar trans-1 2-dimethylcyclohexane Horse conformation judging practice
Horse conformation judging practice Carbohydrates organic chemistry
Carbohydrates organic chemistry Isomerie de conformation
Isomerie de conformation Hsv encephalitis
Hsv encephalitis Roundworms are what type of biohazard virus/parasite
Roundworms are what type of biohazard virus/parasite Recurring decimal questions
Recurring decimal questions It looks for patterns recurring characteristics and events
It looks for patterns recurring characteristics and events Recurring aspects of designs are called design
Recurring aspects of designs are called design Rhythm music appreciation
Rhythm music appreciation Recurring decimal
Recurring decimal The recurring aspects of designs are called design
The recurring aspects of designs are called design The recurring aspects of designs are called design
The recurring aspects of designs are called design Recurring decimal
Recurring decimal Characteristically definition
Characteristically definition Accident chain definition
Accident chain definition Recurring or dominant theme
Recurring or dominant theme Recount the story of pony‘s recurring dream.
Recount the story of pony‘s recurring dream. What is a steady recurring pulse called
What is a steady recurring pulse called What is a steady recurring pulse called
What is a steady recurring pulse called Hình ảnh bộ gõ cơ thể búng tay
Hình ảnh bộ gõ cơ thể búng tay Ng-html
Ng-html Bổ thể
Bổ thể Tỉ lệ cơ thể trẻ em
Tỉ lệ cơ thể trẻ em Gấu đi như thế nào
Gấu đi như thế nào Thang điểm glasgow
Thang điểm glasgow Chúa yêu trần thế alleluia
Chúa yêu trần thế alleluia Các môn thể thao bắt đầu bằng tiếng chạy
Các môn thể thao bắt đầu bằng tiếng chạy