Recap Two ways of using regular expression Search





![Character Classes A character class matches one of the characters in the class: [abc] Character Classes A character class matches one of the characters in the class: [abc]](https://slidetodoc.com/presentation_image_h/0079f6fb76a61a5603593c16b7a321f6/image-8.jpg)







- Slides: 17

Recap: Two ways of using regular expression Search directly: re. search( reg. Exp, text ) 1. Compile reg. Exp to a special format (an SRE_Pattern object) 2. Search for this SRE_Pattern in text 3. Result is an SRE_Match object or precompile the expression: compiled. RE = re. compile( reg. Exp) 1. Now compiled. RE is an SRE_Pattern object compiled. RE. search( text ) 2. Use search method in this SRE_Pattern to search text 3. Result is same SRE_Match object 1

A few more metacharacters ^: indicates placement at the beginning of the string $: indicates placement at the end of the string # search for zero or one t, followed by two a’s # at the beginning of the string: reg. Exp 1 = “^t? aa“ # search for g followed by one or more c’s followed by a # at the end of the string: reg. Exp 1 = “gc+a$“ # whole string should match ct followed by zero or more # g’s followed by a: reg. Exp 1 = “^ctg*a$“ 2

This time we use re. search() to search the text for the regular expressions directly without compiling them in advance Text 1 contains the regular expression ^t? aa Text 1 contains the regular expression gc+a$ Text 2 contains the regular expression ^ctg*a$ 3

Yet more metacharacters. . {}: indicate repetition | : match either regular expression to the left or to the right (): indicate a group (a part of a regular expression) # search for four t’s followed by three c’s: reg. Exp 1 = “t{4}c{3}“ # search for g followed by 1, 2 or 3 c’s: reg. Exp 1 = “gc{1, 3}$“ # search for either gg or cc: reg. Exp 1 = “gg|cc“ # search for either gg or cc followed by tt: reg. Exp 1 = “(gg|cc)tt“ 4

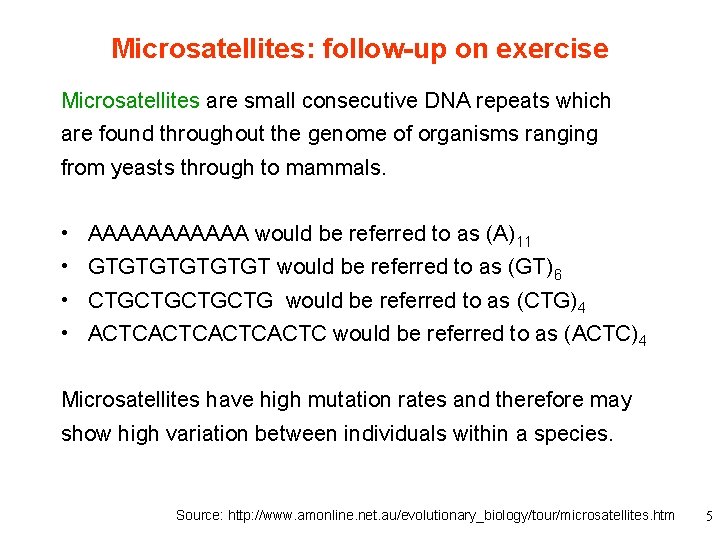
Microsatellites: follow-up on exercise Microsatellites are small consecutive DNA repeats which are found throughout the genome of organisms ranging from yeasts through to mammals. • • AAAAAA would be referred to as (A)11 GTGTGT would be referred to as (GT)6 CTGCTG would be referred to as (CTG)4 ACTCACTC would be referred to as (ACTC)4 Microsatellites have high mutation rates and therefore may show high variation between individuals within a species. Source: http: //www. amonline. net. au/evolutionary_biology/tour/microsatellites. htm 5

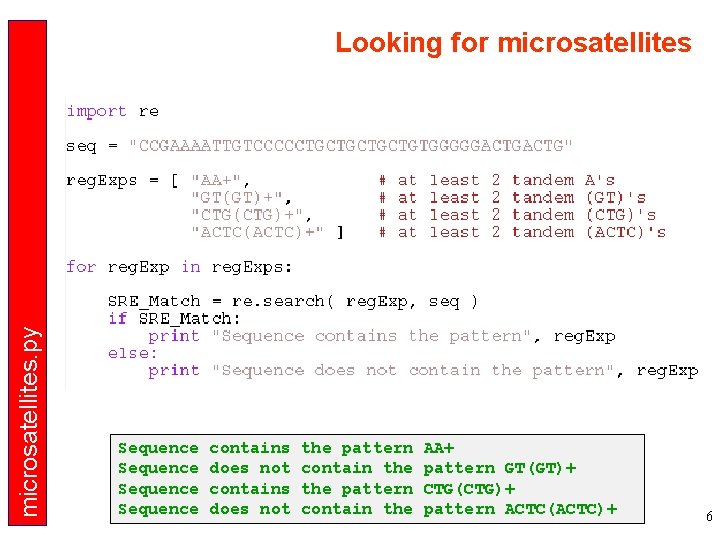
microsatellites. py Looking for microsatellites Sequence contains does not the pattern contain the AA+ pattern GT(GT)+ CTG(CTG)+ pattern ACTC(ACTC)+ 6

Escaping metacharacters : used to escape a metacharacter (“to take it literally”) # search for x followed by + followed by y: reg. Exp 1 = “x+y“ # search for ( followed by x followed by y: reg. Exp 1 = “(xy“ # search for x followed by ? followed by y: reg. Exp 1 = “x? y“ # search for x followed by at least one ^ followed by 3: reg. Exp 1 = “x^+3“ 7
![Character Classes A character class matches one of the characters in the class abc Character Classes A character class matches one of the characters in the class: [abc]](https://slidetodoc.com/presentation_image_h/0079f6fb76a61a5603593c16b7a321f6/image-8.jpg)
Character Classes A character class matches one of the characters in the class: [abc] matches either a or b or c. d[abc]d matches dad and dbd and dcd [ab]+c matches e. g. ac, abc, bac, bbabaabc, . . • Metacharacter ^ at beginning negates character class: [^abc] matches any character other than a, b and c • A class can use – to indicate a range of characters: [a-e] is the same as [abcde] • Characters except ^ and – are taken literally in a class: [a+b*] matches a or + or b or * 8

Special Sequences Special sequence: shortcut for a common character class reg. Exp 1 = “dd: dd [AP]M” # (possibly illegal) time stamps 04: 23: 19 PM reg. Exp 2 = "w+@[w. ]+. dk“ # any Danish email address 9

Regular expression functions sub, split, match reg. Expfunctions. py

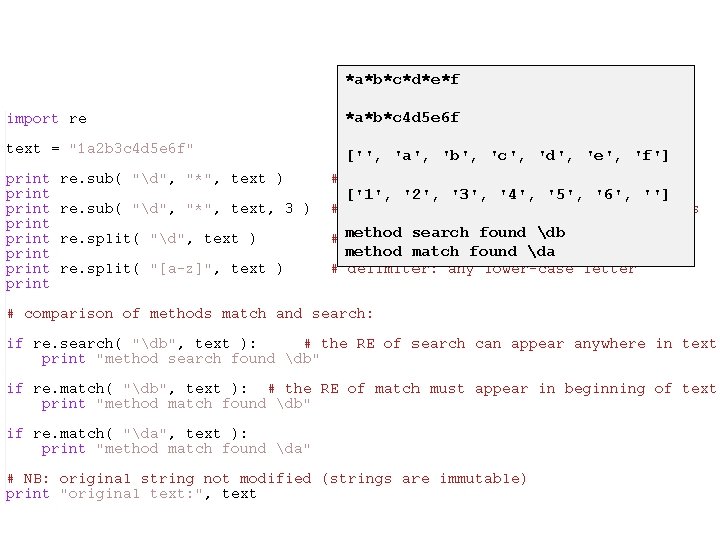
*a*b*c*d*e*f *a*b*c 4 d 5 e 6 f ['', 'a', 'b', 'c', 'd', 'e', 'f'] ['1', '2', '3', '4', '5', '6', ''] method search found db method match found da

If you put ()’s around delimiter pattern, delimiters are returned also reg. Expsplit. py • Recall the trypsin exercise: [‘DCQ’, ‘R’, ‘VYAPFM’, ‘K’, ‘LIHDQWGWDYNNWTSM’, ‘K’, ‘GDA’, ‘R’, ‘EILIMPFCQWTSPF’, ‘R’, ‘NMGCHV’] 12

The group method We can extract the actual substring that matched the regular expression by calling method group() in the SRE_Match object: text = "But here: chili@daimi. au. dk what a *(. @#$ silly @#*. ( email address“ reg. Exp = "w+@[w. ]+. dk“ # match Danish email address compiled. RE = re. compile( reg. Exp) SRE_Match = compiled. RE. search( text ) if SRE_Match: print "Text contains this Danish email address: ", SRE_Match. group() Text contains this Danish email address: chili@daimi. au. dk 13

• • The substring that matches the whole RE is called a group The RE can be subdivided into smaller groups (parts) Each group of the matching substring can be extracted Metacharacters ( and ) denote a group text = "But here: chili@daimi. au. dk what a *(. @#$ silly @#*. ( email address“ # Match any Danish email address; define two groups: username and domain: reg. Exp = “(w+)@([w. ]+. dk)“ compiled. RE = re. compile( reg. Exp ) SRE_Match = compiled. RE. search( text ) if SRE_Match: print "Text contains this Danish email address: ", SRE_Match. group() print “Username: ”, SRE_Match. group(1), “n. Domain: ”, SRE_Match. group(2) danish_emailaddress_groups. py Text contains this Danish email address: chili@daimi. au. dk Username: chili Domain: daimi. au. dk 14

Greedy vs. non-greedy operators • + and * are greedy operators – They attempt to match as many characters as possible • +? and *? are non-greedy operators – They attempt to match as few characters as possible 15

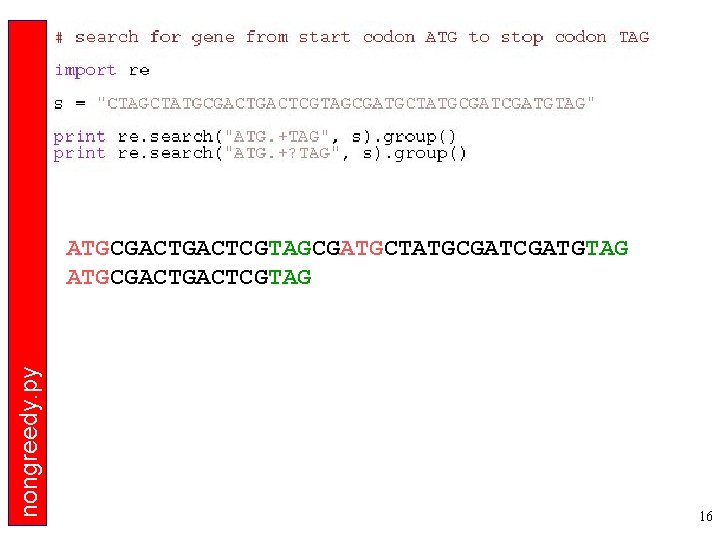
nongreedy. py ATGCGACTCGTAGCGATGCTATGCGATGTAG ATGCGACTCGTAG 16

. . on to the exercises 17