Rational Design Requires 3 D Structure of Enzyme






- Slides: 40

Rational Design • Requires: – 3 D Structure of Enzyme – Knowledge of Enzyme Mechanism • • Increase size of substrate binding pocket Add or subtract an interaction – H-Bond – Ion Pairing – Disulfide Bonds • Replace residues with those found in homologous enzymes • Site-Directed Mutagenesis – Polymerase Chain Reaction Mullis, K. et al. Science 1988, 239, 487

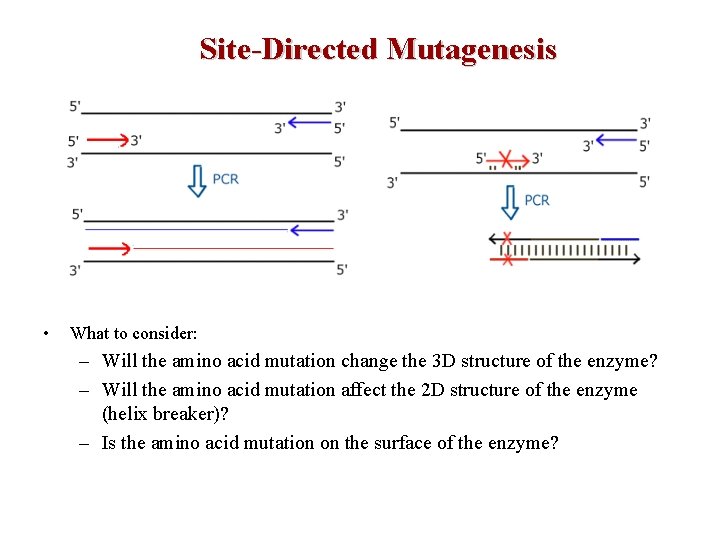
Site-Directed Mutagenesis • What to consider: – Will the amino acid mutation change the 3 D structure of the enzyme? – Will the amino acid mutation affect the 2 D structure of the enzyme (helix breaker)? – Is the amino acid mutation on the surface of the enzyme?

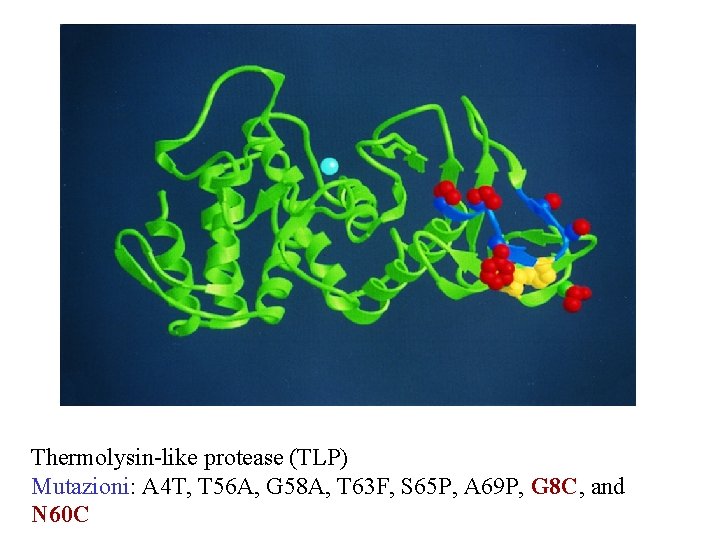
Thermolysin-like protease (TLP) Mutazioni: A 4 T, T 56 A, G 58 A, T 63 F, S 65 P, A 69 P, G 8 C, and N 60 C

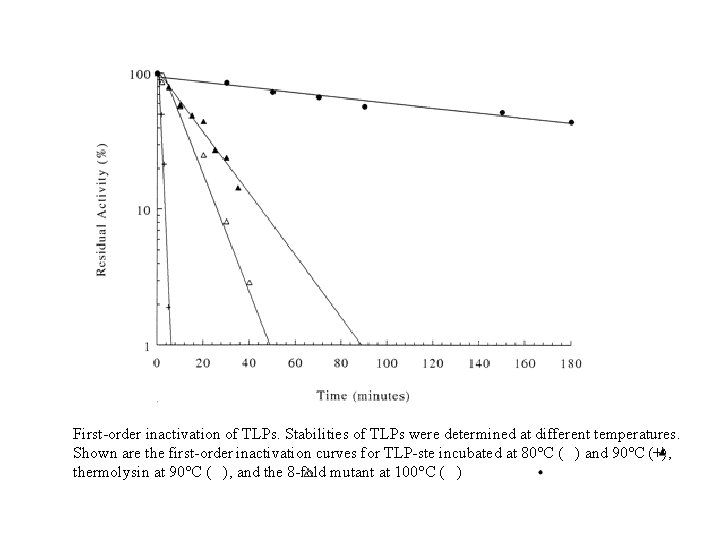
First-order inactivation of TLPs. Stabilities of TLPs were determined at different temperatures. Shown are the first-order inactivation curves for TLP-ste incubated at 80°C ( ) and 90°C (+), thermolysin at 90°C ( ), and the 8 -fold mutant at 100°C ( )

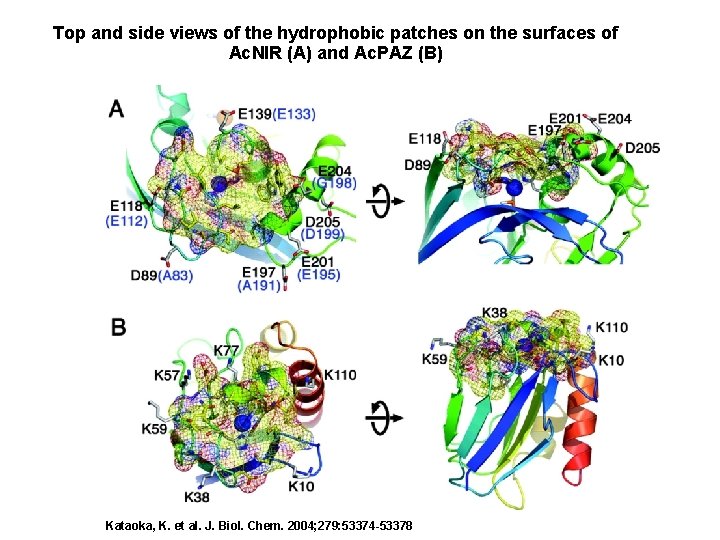
Top and side views of the hydrophobic patches on the surfaces of Ac. NIR (A) and Ac. PAZ (B) Kataoka, K. et al. J. Biol. Chem. 2004; 279: 53374 -53378

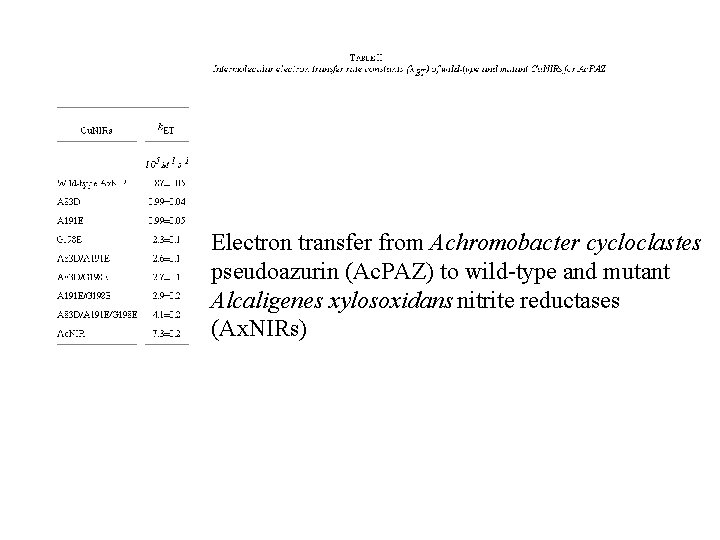
Electron transfer from Achromobacter cycloclastes pseudoazurin (Ac. PAZ) to wild-type and mutant Alcaligenes xylosoxidans nitrite reductases (Ax. NIRs)

“Irrational Design” Directed Evolution

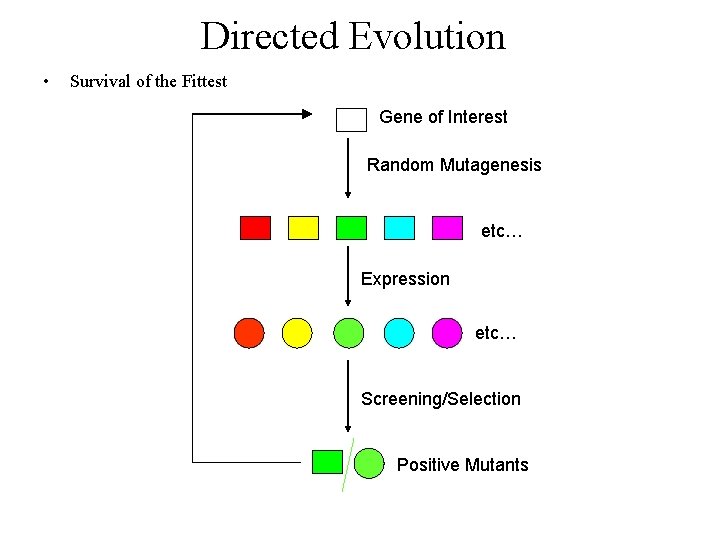
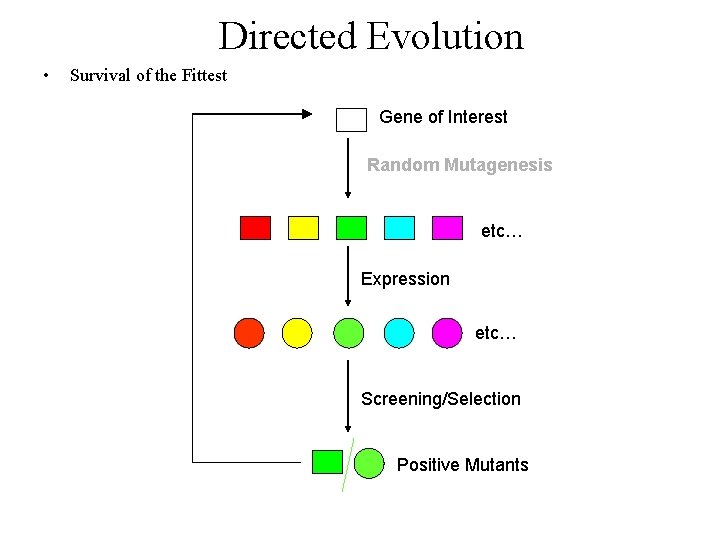
Directed Evolution • Survival of the Fittest Gene of Interest Random Mutagenesis etc… Expression etc… Screening/Selection Positive Mutants

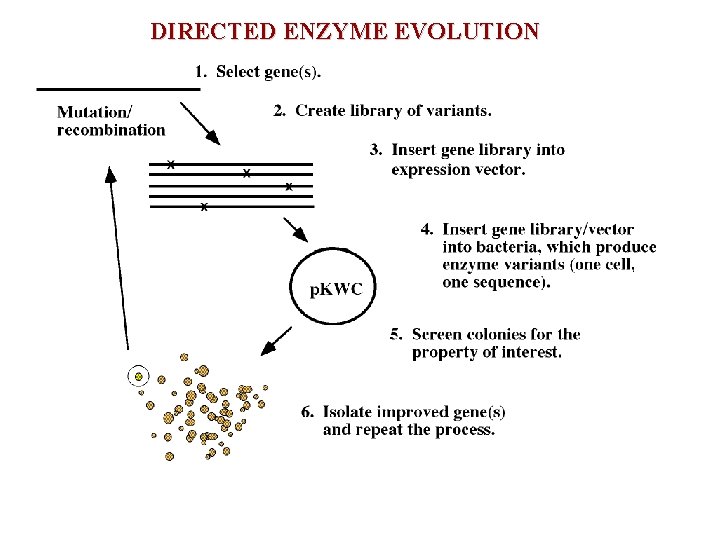
DIRECTED ENZYME EVOLUTION

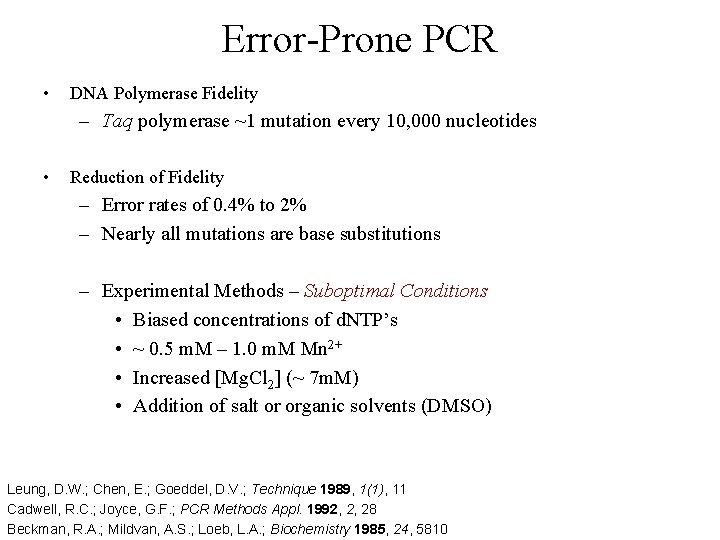
Error-Prone PCR • DNA Polymerase Fidelity – Taq polymerase ~1 mutation every 10, 000 nucleotides • Reduction of Fidelity – Error rates of 0. 4% to 2% – Nearly all mutations are base substitutions – Experimental Methods – Suboptimal Conditions • Biased concentrations of d. NTP’s • ~ 0. 5 m. M – 1. 0 m. M Mn 2+ • Increased [Mg. Cl 2] (~ 7 m. M) • Addition of salt or organic solvents (DMSO) Leung, D. W. ; Chen, E. ; Goeddel, D. V. ; Technique 1989, 1(1), 11 Cadwell, R. C. ; Joyce, G. F. ; PCR Methods Appl. 1992, 2, 28 Beckman, R. A. ; Mildvan, A. S. ; Loeb, L. A. ; Biochemistry 1985, 24, 5810

Error Bias On average only 5. 7 amino acid substitutions are accessible from a 1 base mutation 5’ – AUGUUACAUGGUGCACCACUUAGGCGACCAAAAUGA – 3’ 5’ – AUGUUACAUGGUGCAACACUUAGGCGACCAAAAUGA – 3’ 5’ – AUGUUACAUGGUGCACCUCUUAGGCGACCAAAAUGA – 3’ NH 3+ - Met – Leu – His – Gly – Ala – Pro – Leu – Arg – Leu – Lys – COO NH 3+ - Met – Leu – His – Gly – Ala – Thr – Leu – Arg – Leu – Lys – COONH 3+ - Met – Leu – His – Gly – Ala – Pro – Leu – Arg – Leu – Lys – COO-

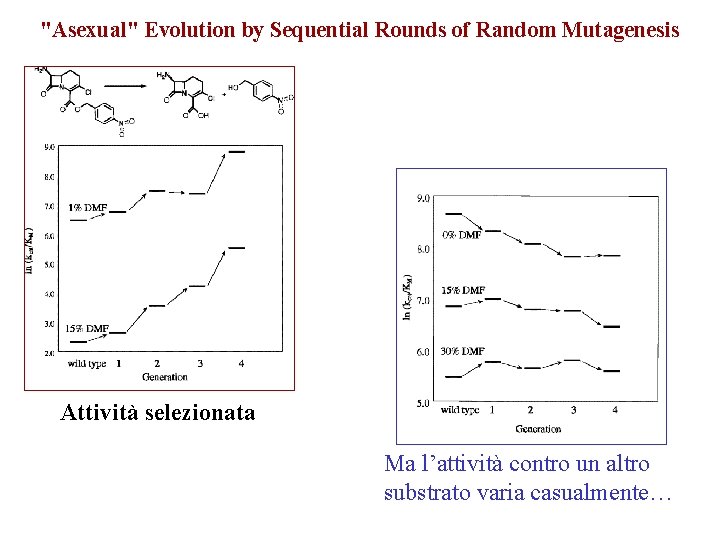
"Asexual" Evolution by Sequential Rounds of Random Mutagenesis Attività selezionata Ma l’attività contro un altro substrato varia casualmente…

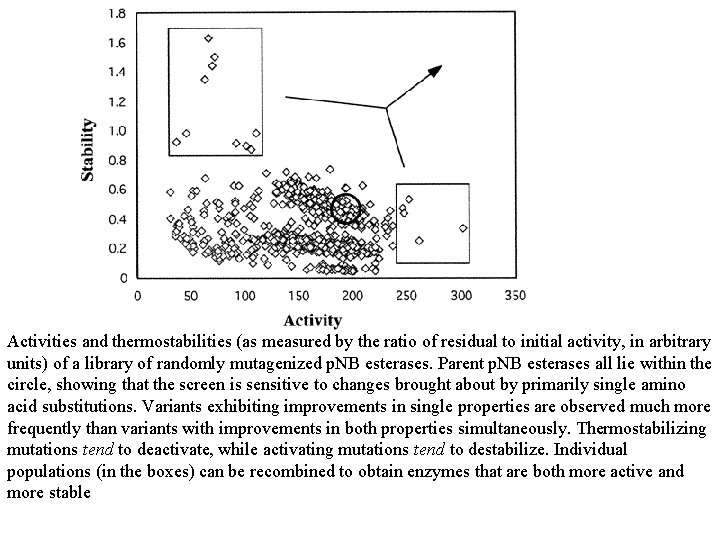
Activities and thermostabilities (as measured by the ratio of residual to initial activity, in arbitrary units) of a library of randomly mutagenized p. NB esterases. Parent p. NB esterases all lie within the circle, showing that the screen is sensitive to changes brought about by primarily single amino acid substitutions. Variants exhibiting improvements in single properties are observed much more frequently than variants with improvements in both properties simultaneously. Thermostabilizing mutations tend to deactivate, while activating mutations tend to destabilize. Individual populations (in the boxes) can be recombined to obtain enzymes that are both more active and more stable

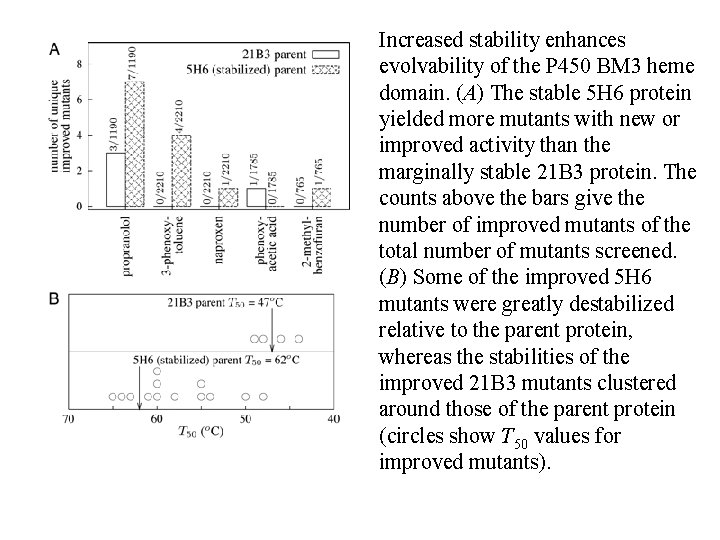
Increased stability enhances evolvability of the P 450 BM 3 heme domain. (A) The stable 5 H 6 protein yielded more mutants with new or improved activity than the marginally stable 21 B 3 protein. The counts above the bars give the number of improved mutants of the total number of mutants screened. (B) Some of the improved 5 H 6 mutants were greatly destabilized relative to the parent protein, whereas the stabilities of the improved 21 B 3 mutants clustered around those of the parent protein (circles show T 50 values for improved mutants).

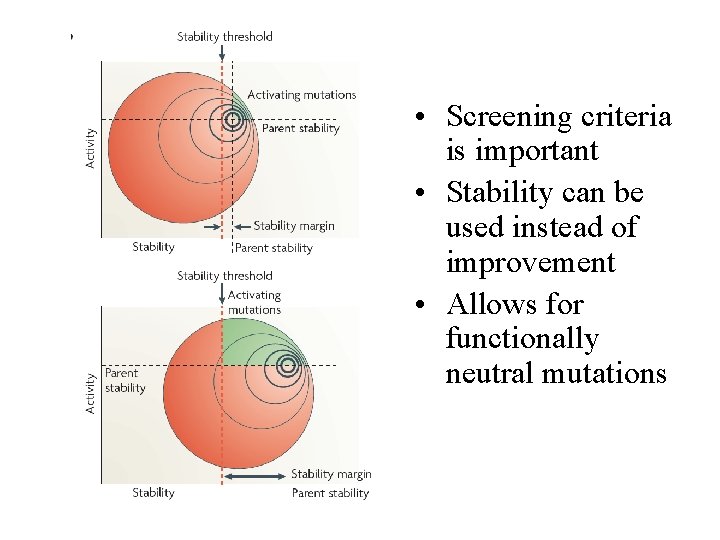
• Screening criteria is important • Stability can be used instead of improvement • Allows for functionally neutral mutations

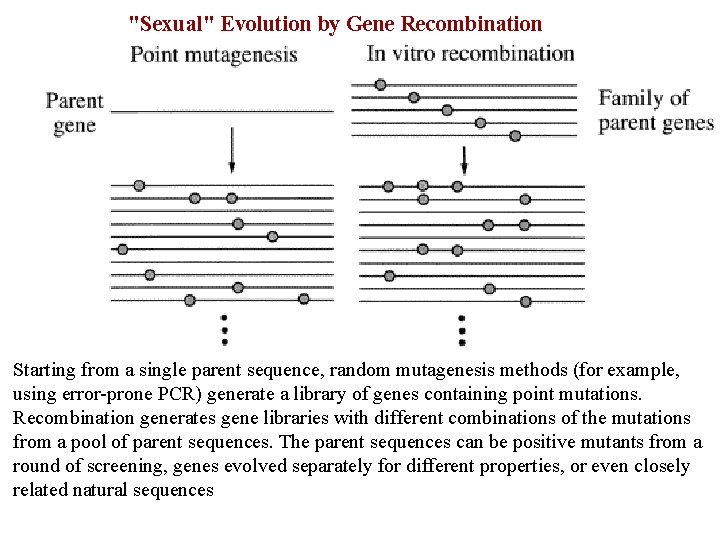
"Sexual" Evolution by Gene Recombination Starting from a single parent sequence, random mutagenesis methods (for example, using error-prone PCR) generate a library of genes containing point mutations. Recombination generates gene libraries with different combinations of the mutations from a pool of parent sequences. The parent sequences can be positive mutants from a round of screening, genes evolved separately for different properties, or even closely related natural sequences

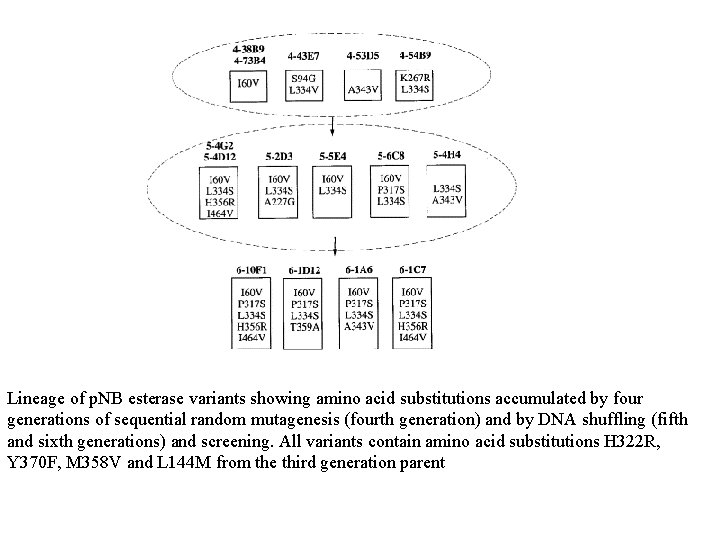
Lineage of p. NB esterase variants showing amino acid substitutions accumulated by four generations of sequential random mutagenesis (fourth generation) and by DNA shuffling (fifth and sixth generations) and screening. All variants contain amino acid substitutions H 322 R, Y 370 F, M 358 V and L 144 M from the third generation parent

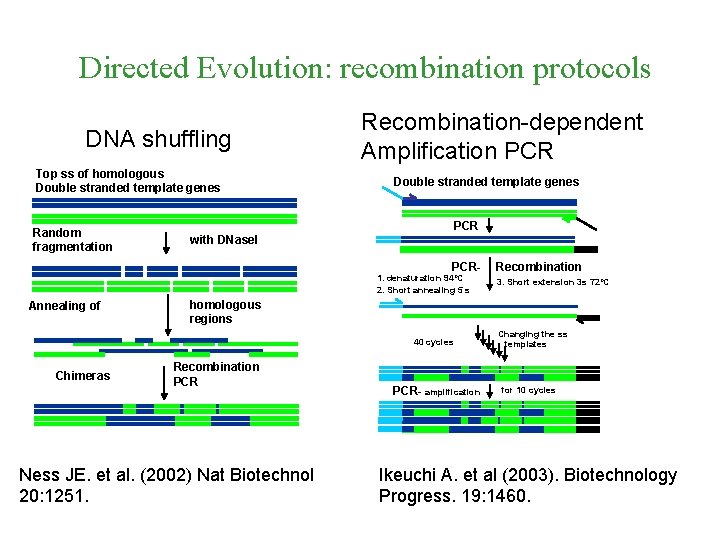
Directed Evolution: recombination protocols DNA shuffling Top ss of homologous Double stranded template genes Random fragmentation Recombination-dependent Amplification PCR Double stranded template genes PCR with DNase. I PCR 1. denaturation 94ºC 2. Short annealing 5 s Annealing of 3. Short extension 3 s 72ºC homologous regions 40 cycles Chimeras Recombination PCR Ness JE. et al. (2002) Nat Biotechnol 20: 1251. PCR- amplification Changing the ss templates for 10 cycles Ikeuchi A. et al (2003). Biotechnology Progress. 19: 1460.

Directed Evolution • Survival of the Fittest Gene of Interest Random Mutagenesis etc… Expression etc… Screening/Selection Positive Mutants

Screening • High-Throughput Methods • Selection – UV/Vis Spectroscopy – IR Spectroscopy – Capillary Array Electrophoresis – GC – p. H indicators – Fluorescence – Circular Dichroism – Mass Spectrometry Angew. Chem. Int. Ed. Engl. 1997, 36, 2830 Angew. Chem. Int. Ed. Engl. 1998, 37, 2647 Angew. Chem. Int. Ed. Engl. 2000, 39, 3891 Catal. Today 2001, 67, 389 • Phage Display Chem. Eur. J. 1998, 4, 2324 Angew. Chem. Int. Ed. Engl. 1999, 38, 497 Angew. Chem. Int. Ed. Engl. 1999, 111, 1868

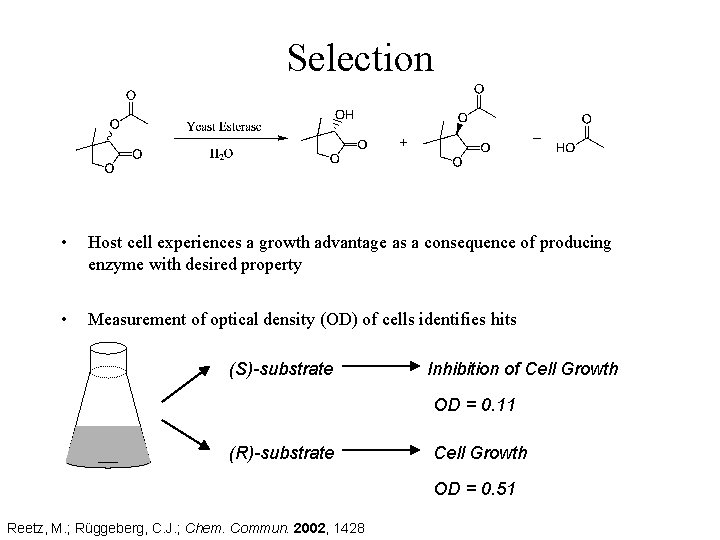
Selection • Host cell experiences a growth advantage as a consequence of producing enzyme with desired property • Measurement of optical density (OD) of cells identifies hits (S)-substrate Inhibition of Cell Growth OD = 0. 11 (R)-substrate Cell Growth OD = 0. 51 Reetz, M. ; Rüggeberg, C. J. ; Chem. Commun. 2002, 1428

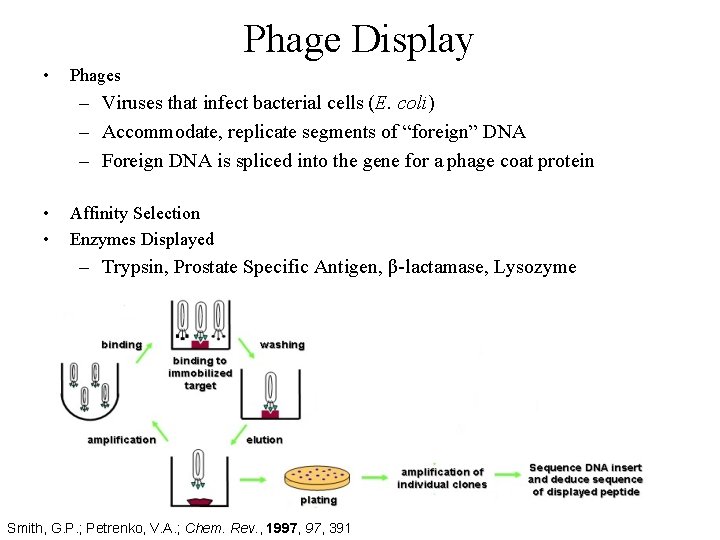
Phage Display • Phages – Viruses that infect bacterial cells (E. coli) – Accommodate, replicate segments of “foreign” DNA – Foreign DNA is spliced into the gene for a phage coat protein • • Affinity Selection Enzymes Displayed – Trypsin, Prostate Specific Antigen, β-lactamase, Lysozyme Smith, G. P. ; Petrenko, V. A. ; Chem. Rev. , 1997, 391

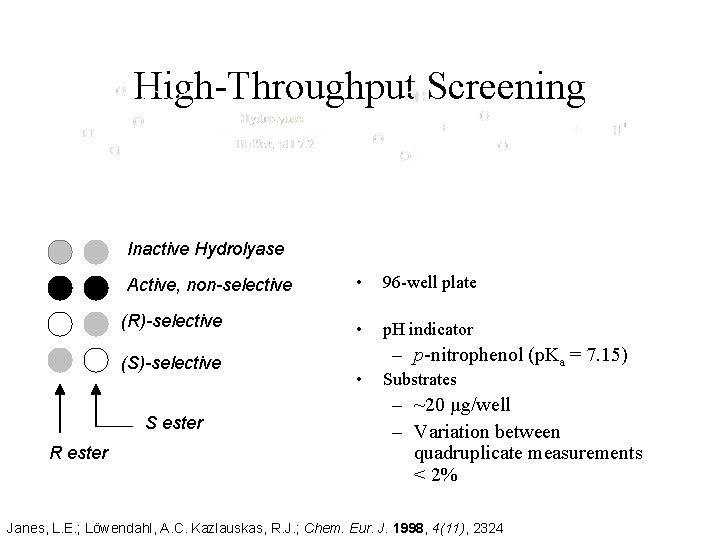
High-Throughput Screening Inactive Hydrolyase Active, non-selective (R)-selective (S)-selective S ester R ester • 96 -well plate • p. H indicator • – p-nitrophenol (p. Ka = 7. 15) Substrates – ~20 μg/well – Variation between quadruplicate measurements < 2% Janes, L. E. ; Löwendahl, A. C. Kazlauskas, R. J. ; Chem. Eur. J. 1998, 4(11), 2324

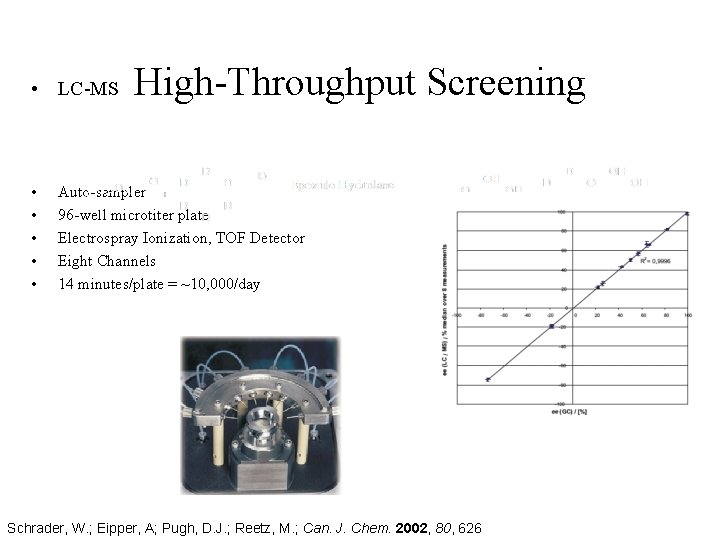
• LC-MS • • • High-Throughput Screening Auto-sampler 96 -well microtiter plate Electrospray Ionization, TOF Detector Eight Channels 14 minutes/plate = ~10, 000/day Schrader, W. ; Eipper, A; Pugh, D. J. ; Reetz, M. ; Can. J. Chem. 2002, 80, 626


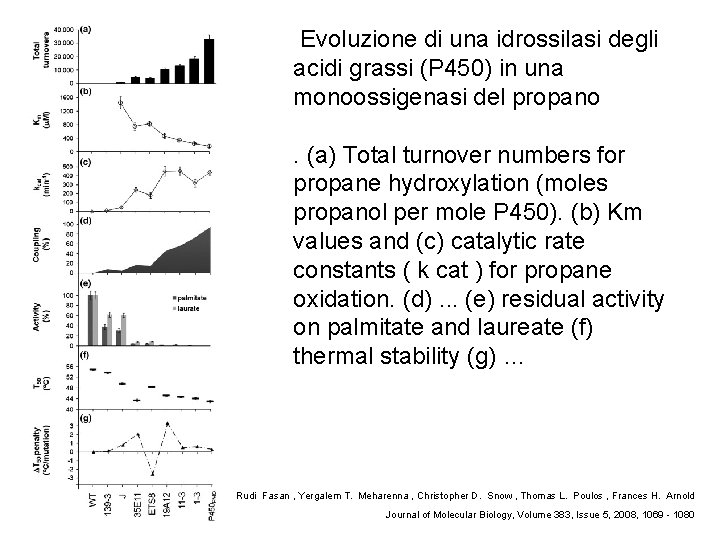
Evoluzione di una idrossilasi degli acidi grassi (P 450) in una monoossigenasi del propano. (a) Total turnover numbers for propane hydroxylation (moles propanol per mole P 450). (b) Km values and (c) catalytic rate constants ( k cat ) for propane oxidation. (d). . . (e) residual activity on palmitate and laureate (f) thermal stability (g) … Rudi Fasan , Yergalem T. Meharenna , Christopher D. Snow , Thomas L. Poulos , Frances H. Arnold Journal of Molecular Biology, Volume 383, Issue 5, 2008, 1069 - 1080

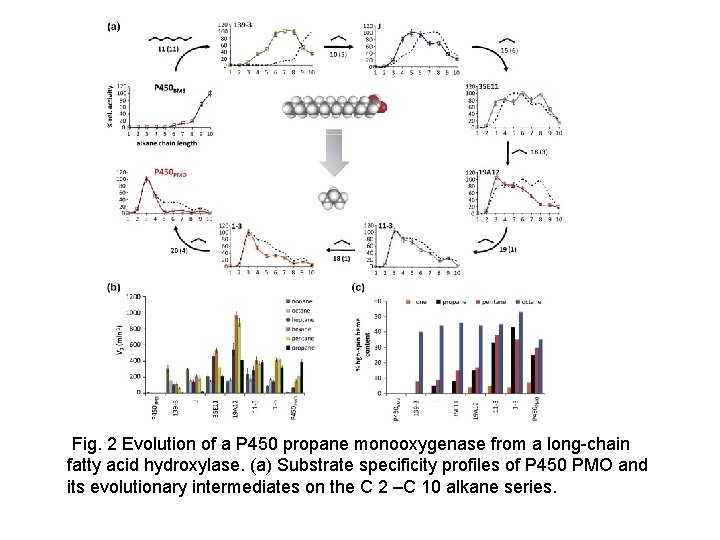
Fig. 2 Evolution of a P 450 propane monooxygenase from a long-chain fatty acid hydroxylase. (a) Substrate specificity profiles of P 450 PMO and its evolutionary intermediates on the C 2 –C 10 alkane series.

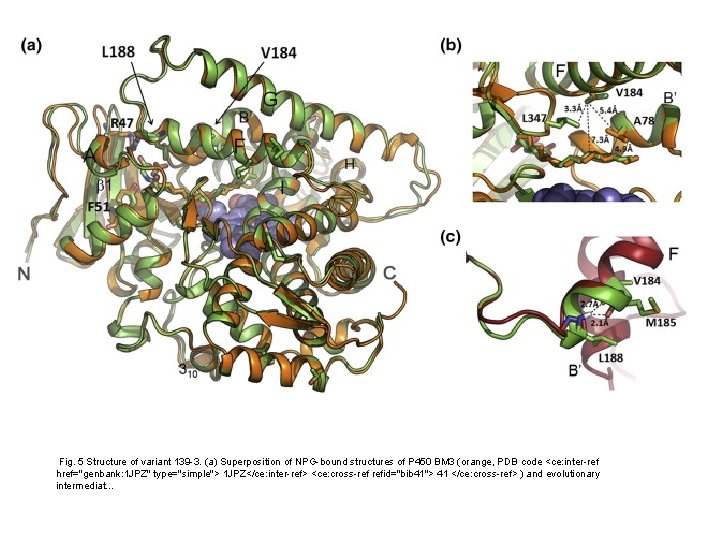
Fig. 5 Structure of variant 139 -3. (a) Superposition of NPG-bound structures of P 450 BM 3 (orange, PDB code <ce: inter-ref href="genbank: 1 JPZ" type="simple"> 1 JPZ</ce: inter-ref> <ce: cross-ref refid="bib 41"> 41 </ce: cross-ref> ) and evolutionary intermediat. . .

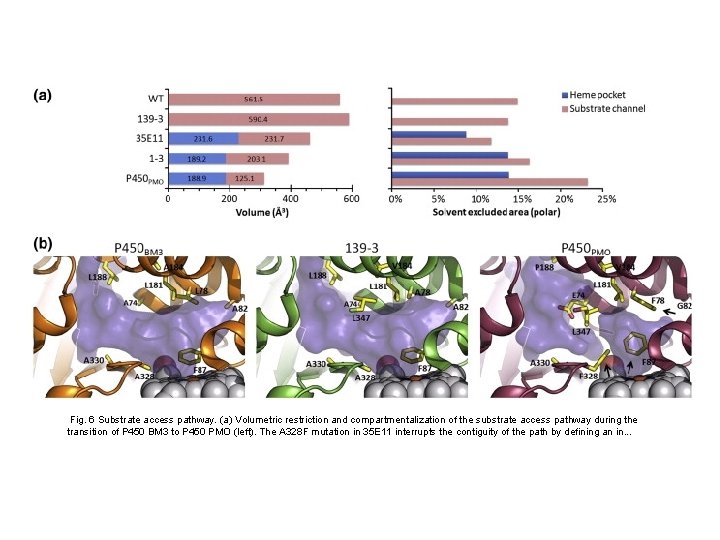
Fig. 6 Substrate access pathway. (a) Volumetric restriction and compartmentalization of the substrate access pathway during the transition of P 450 BM 3 to P 450 PMO (left). The A 328 F mutation in 35 E 11 interrupts the contiguity of the path by defining an in. . .

Toluene o. Monooxygenase or “TOM” • Polymer of 1500 units (amino acids) • At each position there are 19 possibilities (20 building blocks, 1 already used) • 191500 possibilities (101918)

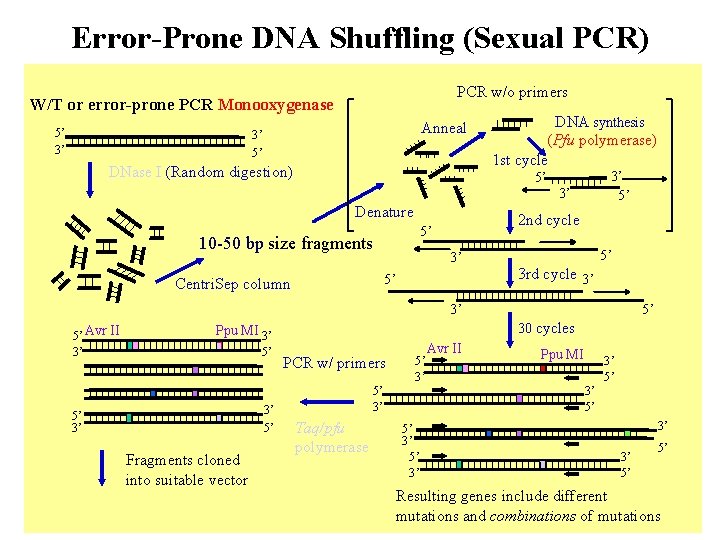
Error-Prone DNA Shuffling (Sexual PCR) PCR w/o primers W/T or error-prone PCR Monooxygenase 5’ 3’ Anneal 3’ 5’ DNase I (Random digestion) DNA synthesis (Pfu polymerase) 1 st cycle 5’ Denature 3’ 5’ Centri. Sep column 3’ 2 nd cycle 5’ 10 -50 bp size fragments 3’ 5’ 5’ 3 rd cycle 3’ 3’ 5’ Avr II 3’ 30 cycles Ppu MI 3’ 5’ 5’ 3’ Fragments cloned into suitable vector 5’ PCR w/ primers 5’ 3’ Taq/pfu polymerase 5’ 3’ Avr II Ppu MI 3’ 5’ 3’ 3’ 5’ 5’ Resulting genes include different mutations and combinations of mutations

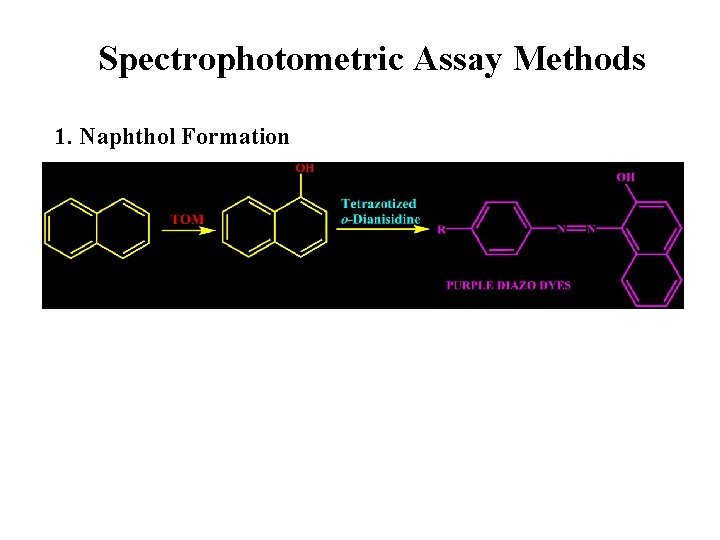
Spectrophotometric Assay Methods 1. Naphthol Formation

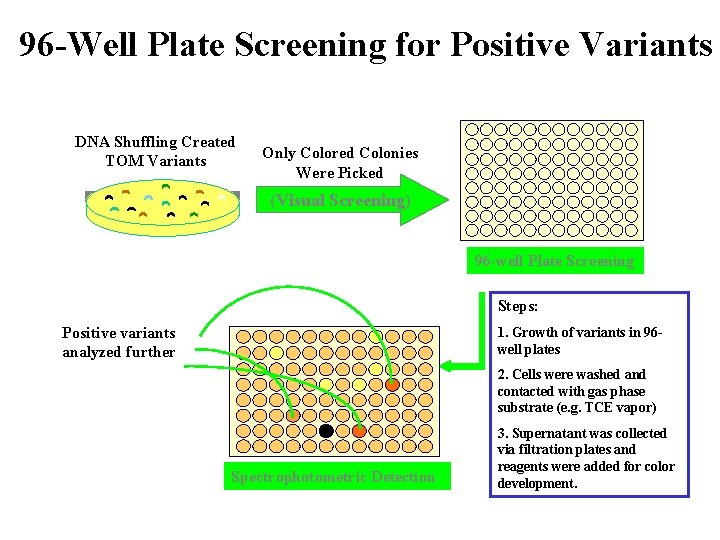
96 -Well Plate Screening for Positive Variants DNA Shuffling Created TOM Variants Only Colored Colonies Were Picked (Visual Screening) 96 -well Plate Screening Steps: Positive variants analyzed further 1. Growth of variants in 96 well plates 2. Cells were washed and contacted with gas phase substrate (e. g. TCE vapor) Spectrophotometric Detection 3. Supernatant was collected via filtration plates and reagents were added for color development.

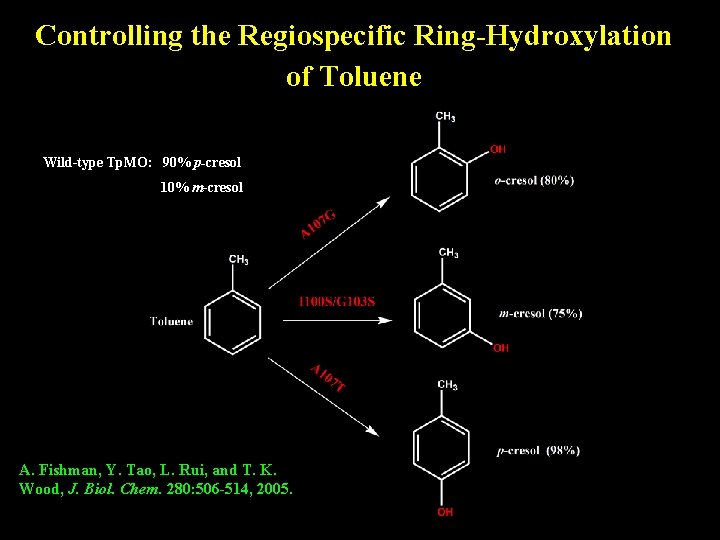
Controlling the Regiospecific Ring-Hydroxylation of Toluene Wild-type Tp. MO: 90% p-cresol 10% m-cresol A. Fishman, Y. Tao, L. Rui, and T. K. Wood, J. Biol. Chem. 280: 506 -514, 2005.

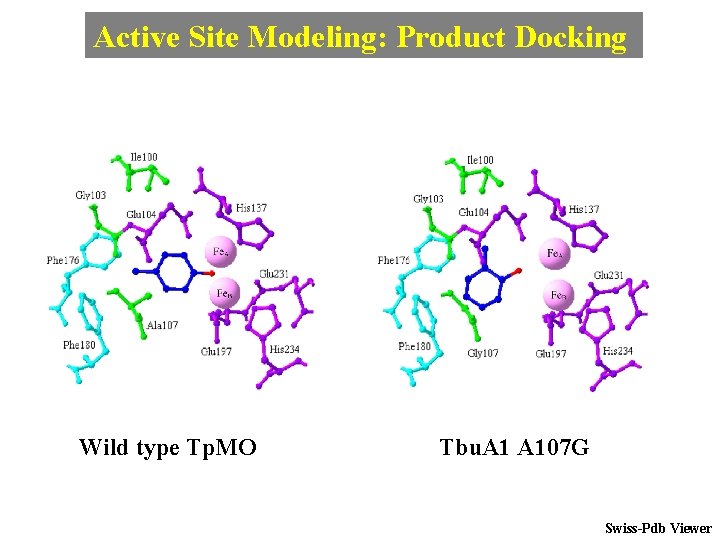
Active Site Modeling: Product Docking Wild type Tp. MO Tbu. A 107 G Swiss-Pdb Viewer

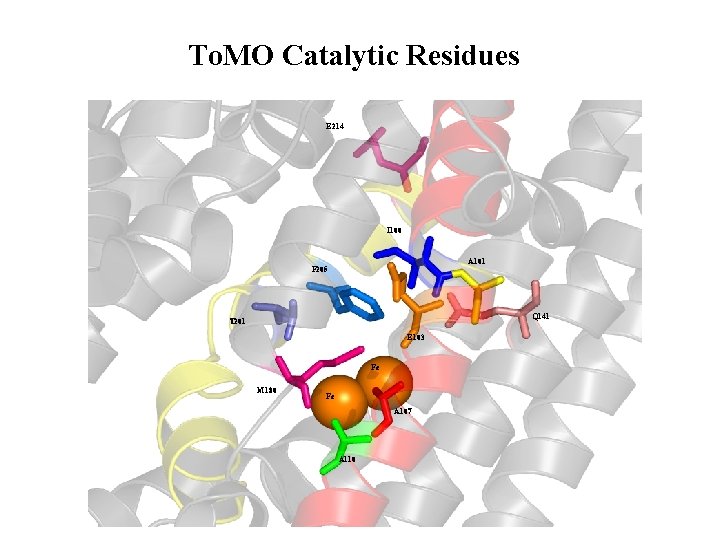
To. MO Catalytic Residues E 214 I 100 A 101 F 205 Q 141 T 201 E 103 Fe M 180 Fe A 107 A 110

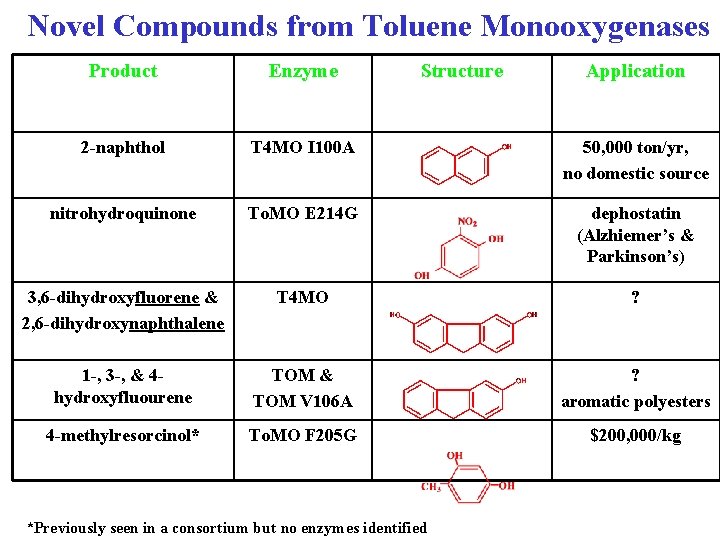
Novel Compounds from Toluene Monooxygenases Product Enzyme Structure 2 -naphthol T 4 MO I 100 A 50, 000 ton/yr, no domestic source nitrohydroquinone To. MO E 214 G dephostatin (Alzhiemer’s & Parkinson’s) 3, 6 -dihydroxyfluorene & 2, 6 -dihydroxynaphthalene T 4 MO ? 1 -, 3 -, & 4 hydroxyfluourene TOM & TOM V 106 A ? aromatic polyesters 4 -methylresorcinol* To. MO F 205 G $200, 000/kg *Previously seen in a consortium but no enzymes identified Application

Color Chemistry

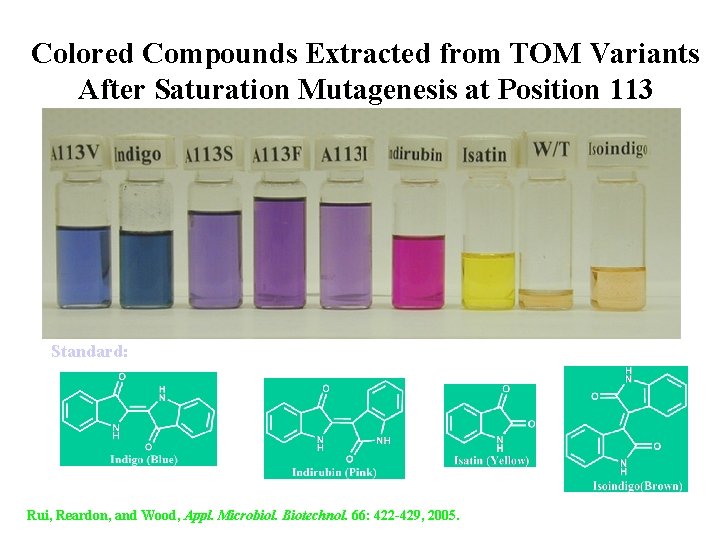
Colored Compounds Extracted from TOM Variants After Saturation Mutagenesis at Position 113 Standard: Rui, Reardon, and Wood, Appl. Microbiol. Biotechnol. 66: 422 -429, 2005.

Conclusioni • Il DNA shuffling può essere usato per individuare quali amino acidi sono coinvolti nella catalisi • La mutagenesi della Tp. MO alle posizioni I 100, G 103, e A 107 permette di modificare la regiochimica dell’ossidazione di toluene e naftalene, ottenendo anche il meta-cresolo, che gli enzimi naturali non producono • La mutazione della A 113 nella toluene monossigenasi modifica la regiospecificità dell’ossidazione dell’indolo, generando diversi colori • Uso come catalizzatori per chimica fine