Rapid Dereplication using Capillary NMR and a Database

Rapid Dereplication using Capillary NMR and a Database of Structures John Blunt University of Canterbury, NZ (A presentation given at the Gordon Research Conference on Marine Natural Products in Ventura, CA, February 2006. Several of the concepts contained herein had been presented earlier at other conferences and institutions during 2003 -2005. )

Acknowledgements Development of the concept and techniques for the use of HPLC-microtitre plate-capillary NMR: John Blunt & Murray Munro (UC) Kirk Gustafson (MTDP, NCI, Frederick MD) Development of the concept of and construction of databases for use in dereplication: John Blunt & Murray Munro (UC) Hartmut Laatsch (University of Goettingen) Preparation of samples for demonstration of techniques: Gill Ellis, Gerhard Lang, Jackson Sun Lin, Maya Mitova Richard Phipps & Sonia van der Sar (UC)

Dereplication Determining which of the bioactive components in an extract are known compounds, or are likely to be new and therefore worthy of further attention.
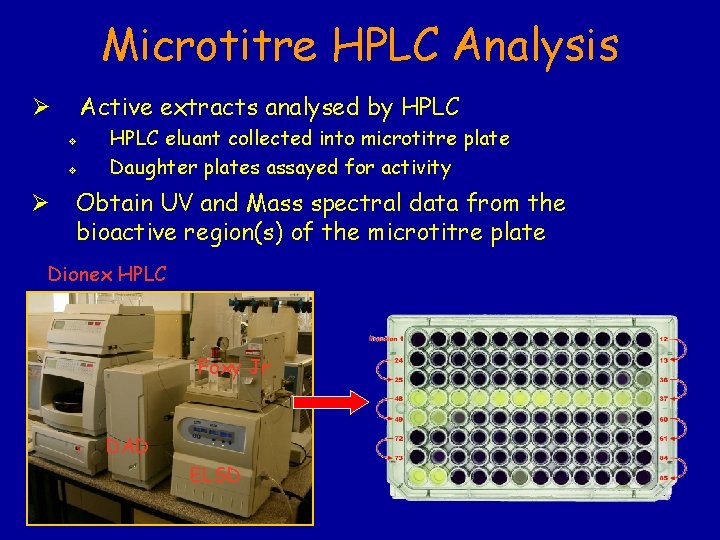
Microtitre HPLC Analysis Active extracts analysed by HPLC Ø v v Ø HPLC eluant collected into microtitre plate Daughter plates assayed for activity Obtain UV and Mass spectral data from the bioactive region(s) of the microtitre plate Dionex HPLC Foxy Jr DAD ELSD
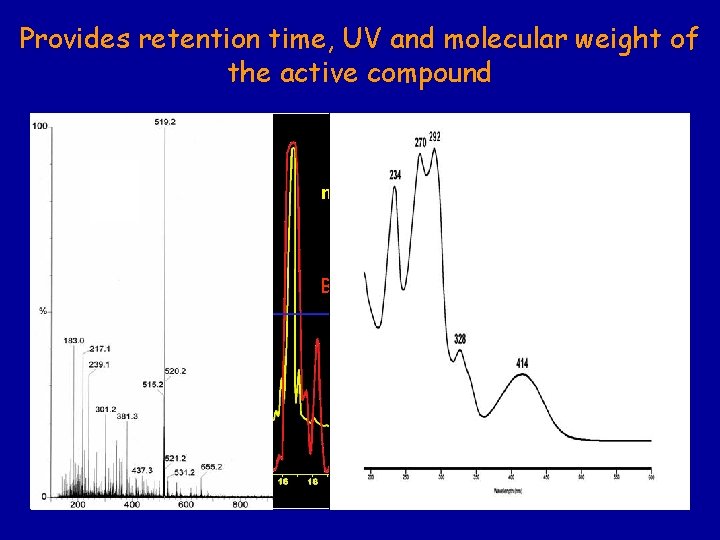
Provides retention time, UV and molecular weight of the active compound C 18 HPLC of extract from marine-derived Cephalosporium sp. DAD detection Bioactivity profile
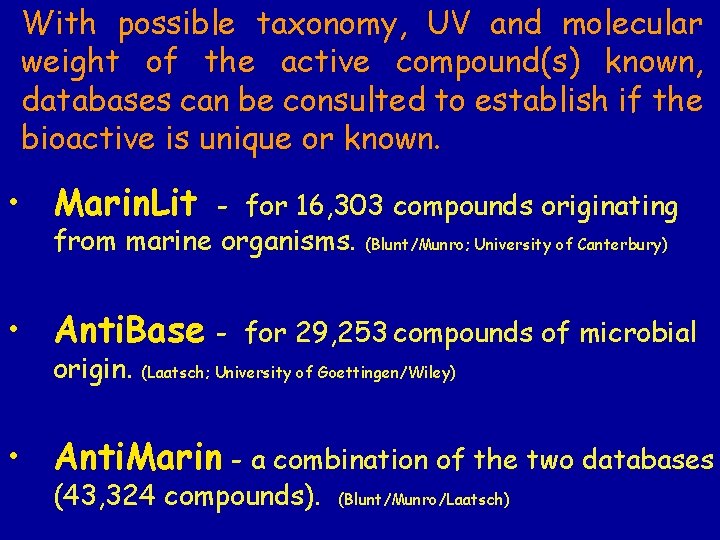
With possible taxonomy, UV and molecular weight of the active compound(s) known, databases can be consulted to establish if the bioactive is unique or known. • Marin. Lit - for 16, 303 compounds originating from marine organisms. (Blunt/Munro; University of Canterbury) • Anti. Base origin. for 29, 253 compounds of microbial (Laatsch; University of Goettingen/Wiley) • Anti. Marin - a combination of the two databases (43, 324 compounds). (Blunt/Munro/Laatsch)
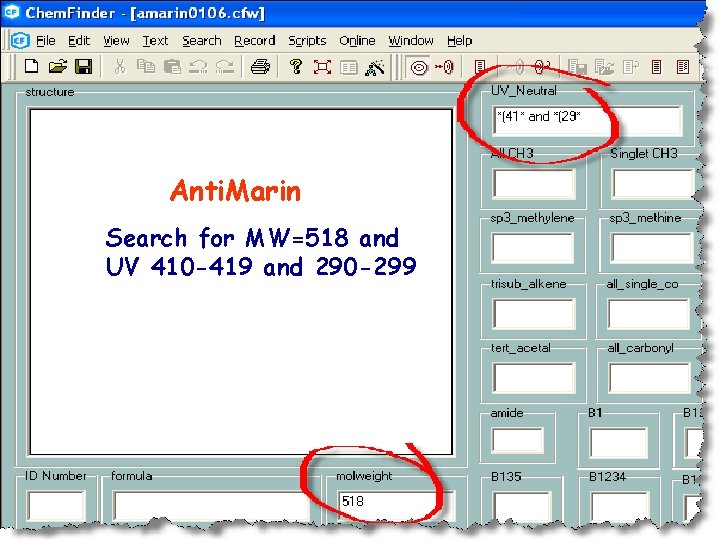
Anti. Marin Search for MW=518 and UV 410 -419 and 290 -299
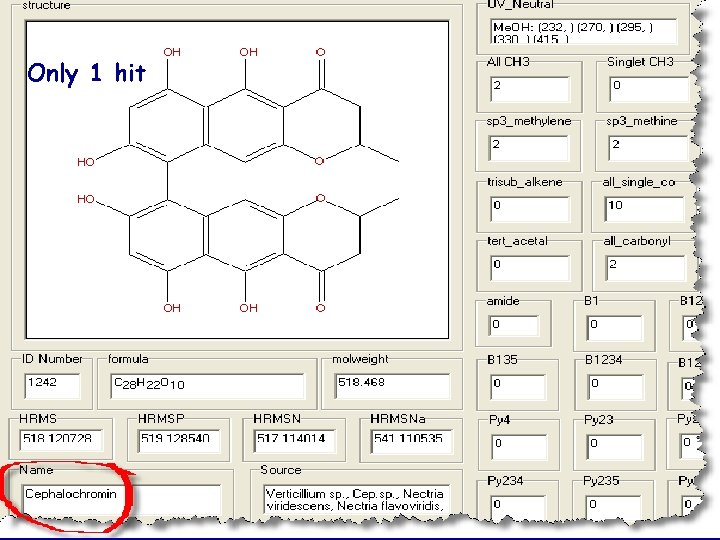
Only 1 hit
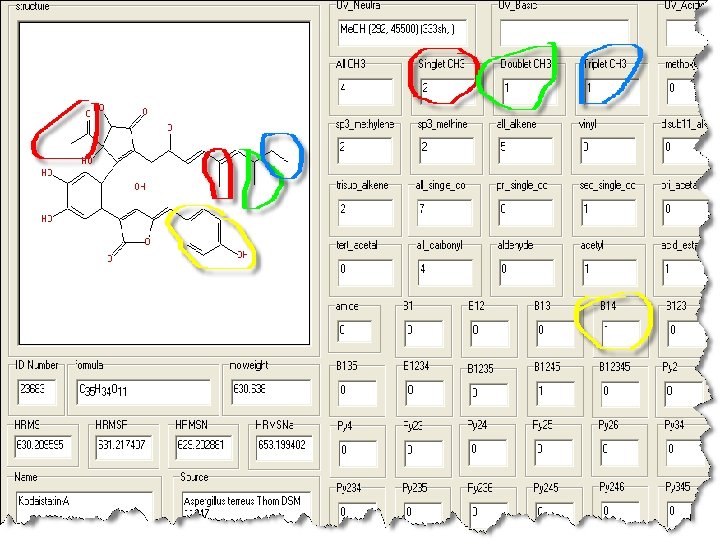
• Problem – not much UV data in Anti. Base • Solution – use 1 H NMR data All structures in Anti. Marin are coded with the numbers of each structural feature that could be deduced from 1 H NMR spectra eg #s of CH 3 of different types (s, d, t)
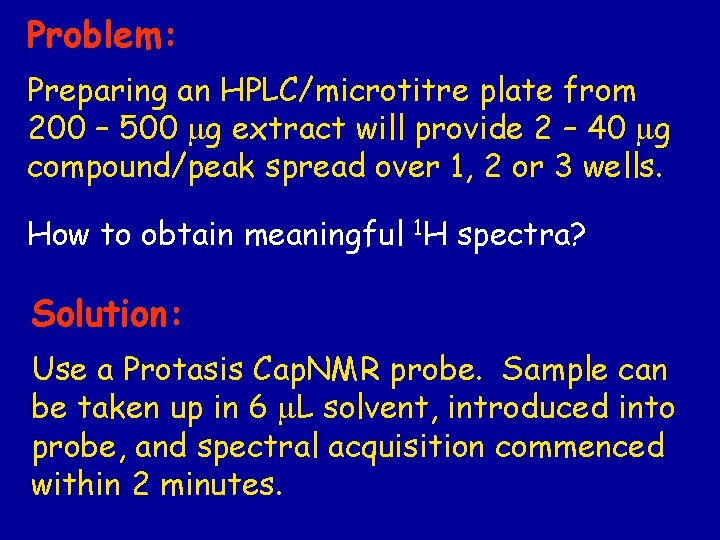
Problem: Preparing an HPLC/microtitre plate from 200 – 500 mg extract will provide 2 – 40 mg compound/peak spread over 1, 2 or 3 wells. How to obtain meaningful 1 H spectra? Solution: Use a Protasis Cap. NMR probe. Sample can be taken up in 6 m. L solvent, introduced into probe, and spectral acquisition commenced within 2 minutes.

Protasis Cap. NMR Probe

HPLC with ELSD detection of 600 mg extract of unidentified endophytic fungus E 9, E 10, E 11 250 m. L/well G 4
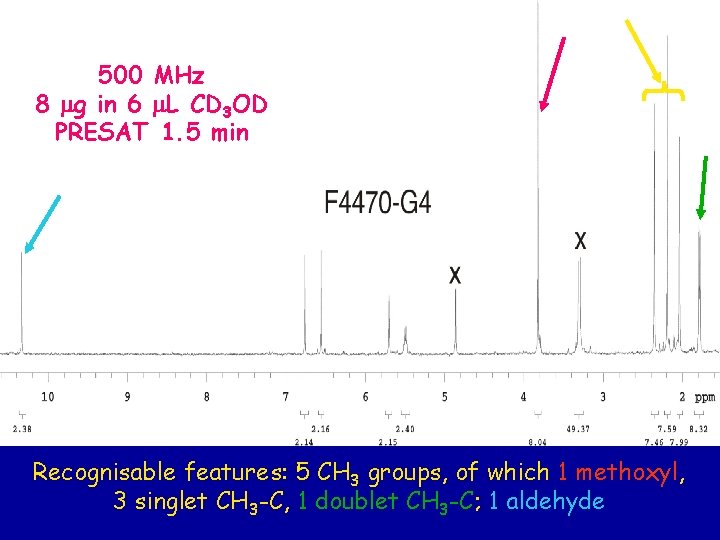
500 MHz 8 mg in 6 m. L CD 3 OD PRESAT 1. 5 min Recognisable features: 5 CH 3 groups, of which 1 methoxyl, 3 singlet CH 3 -C, 1 doublet CH 3 -C; 1 aldehyde

Anti. Marin search for all compounds containing 1 aldehyde and 5 methyl groups, of which 1 is methoxyl, 3 are singlet CH 3 -C, and 1 is doublet CH 3 -C
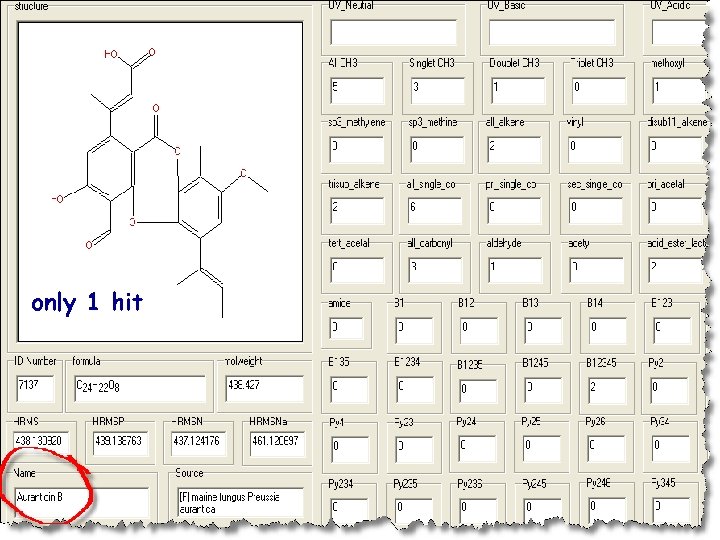
only 1 hit
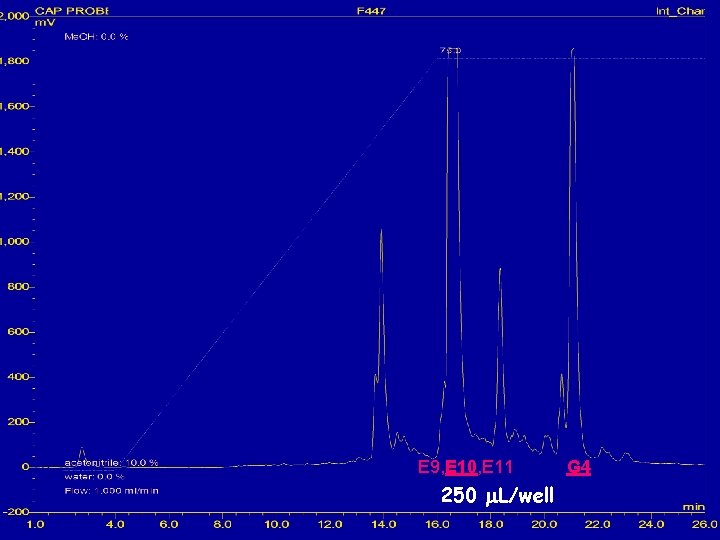
E 9, E 10, E 11 250 m. L/well G 4
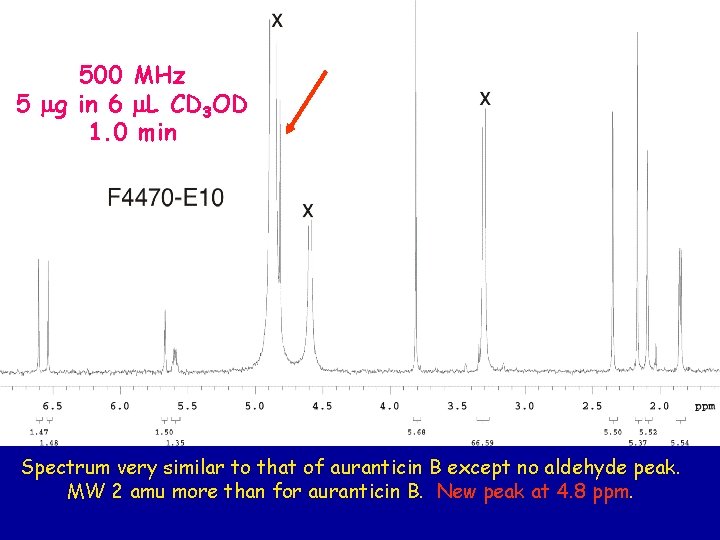
500 MHz 5 mg in 6 m. L CD 3 OD 1. 0 min Spectrum very similar to that of auranticin B except no aldehyde peak. MW 2 amu more than for auranticin B. New peak at 4. 8 ppm.
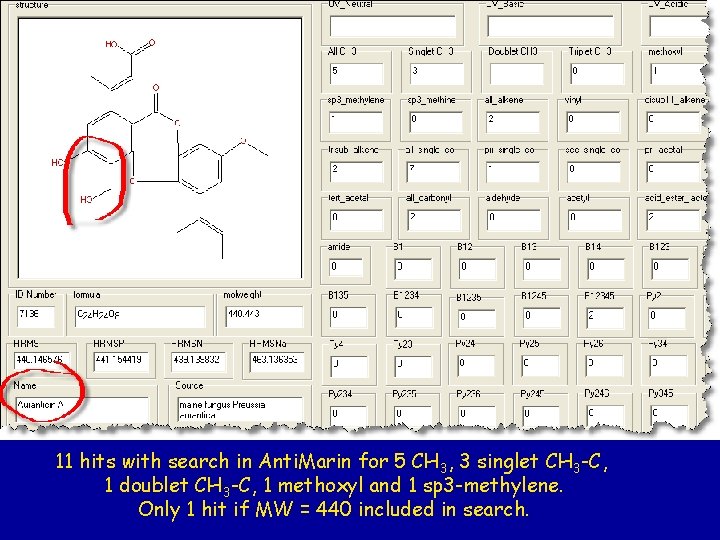
11 hits with search in Anti. Marin for 5 CH 3, 3 singlet CH 3 -C, 1 doublet CH 3 -C, 1 methoxyl and 1 sp 3 -methylene. Only 1 hit if MW = 440 included in search.
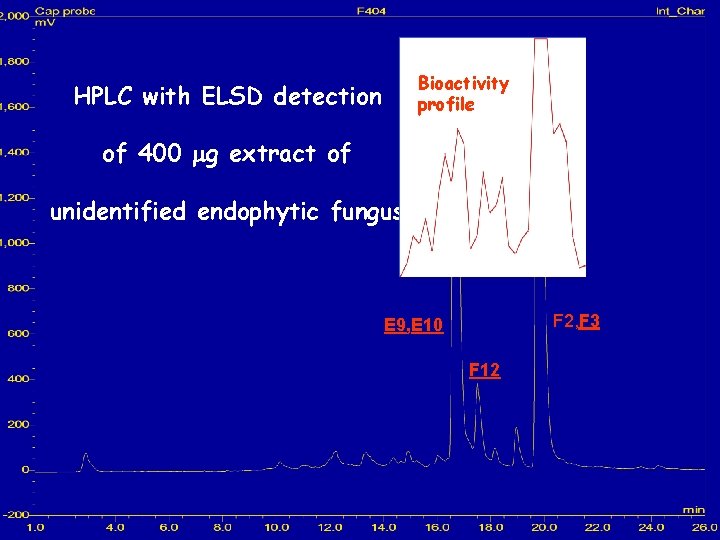
Bioactivity profile HPLC with ELSD detection of 400 mg extract of unidentified endophytic fungus F 2, F 3 E 9, E 10 F 12
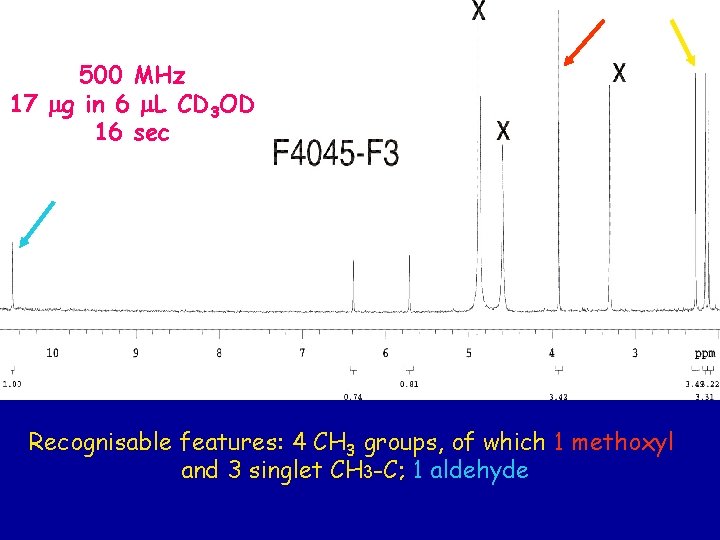
500 MHz 17 mg in 6 m. L CD 3 OD 16 sec Recognisable features: 4 CH 3 groups, of which 1 methoxyl and 3 singlet CH 3 -C; 1 aldehyde
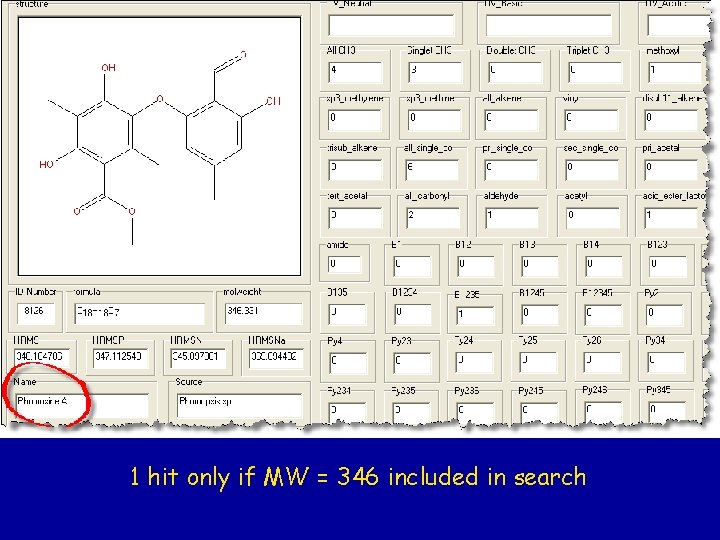
1 hit only if MW = 346 included in search
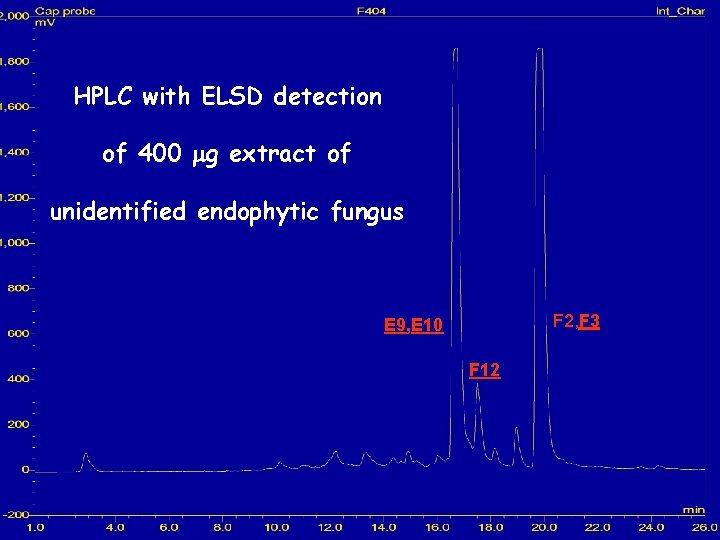
HPLC with ELSD detection of 400 mg extract of unidentified endophytic fungus F 2, F 3 E 9, E 10 F 12
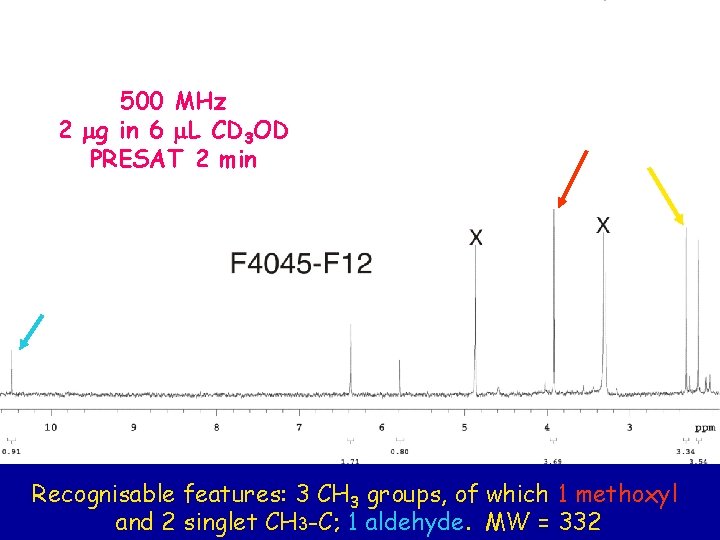
500 MHz 2 mg in 6 m. L CD 3 OD PRESAT 2 min Recognisable features: 3 CH 3 groups, of which 1 methoxyl and 2 singlet CH 3 -C; 1 aldehyde. MW = 332
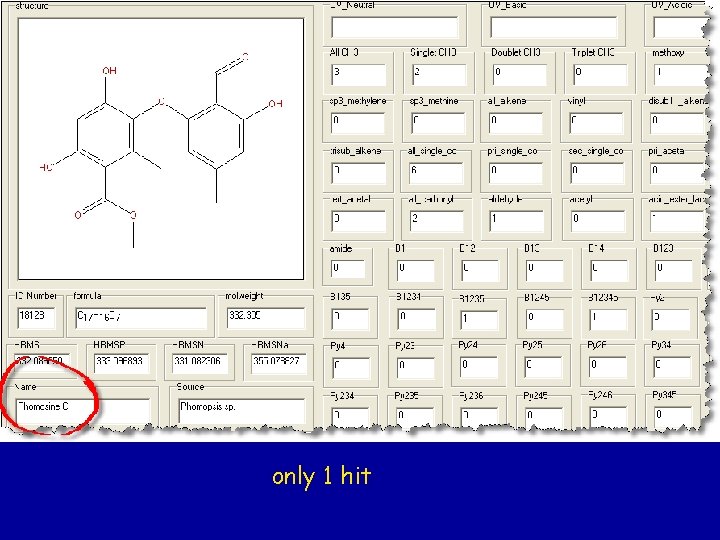
only 1 hit
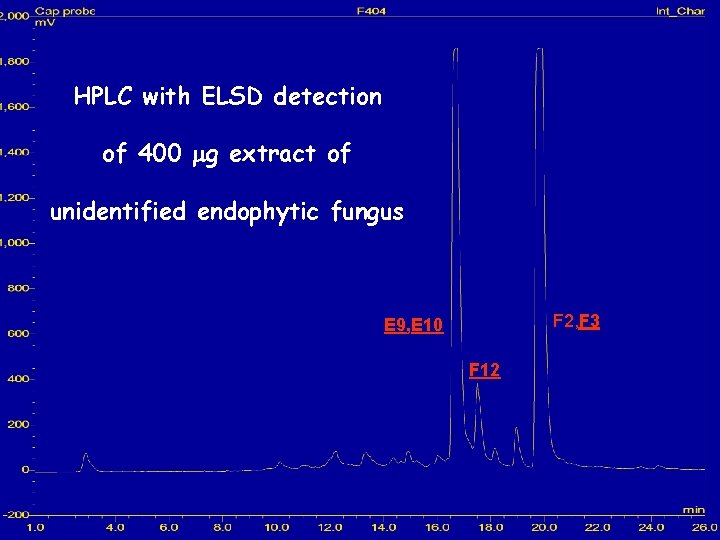
HPLC with ELSD detection of 400 mg extract of unidentified endophytic fungus F 2, F 3 E 9, E 10 F 12
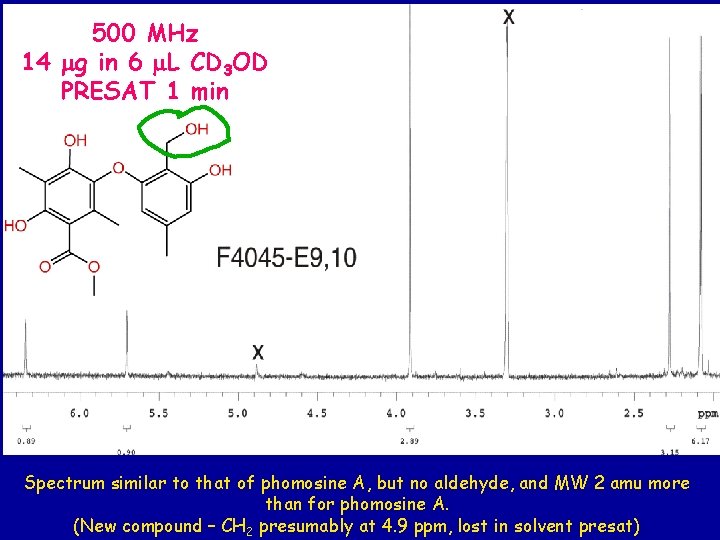
500 MHz 14 mg in 6 m. L CD 3 OD PRESAT 1 min Spectrum similar to that of phomosine A, but no aldehyde, and MW 2 amu more than for phomosine A. (New compound – CH 2 presumably at 4. 9 ppm, lost in solvent presat)

HPLC with ELSD detection of 500 mg extract of unidentified endophytic fungus F 2, F 3, F 4 H 11
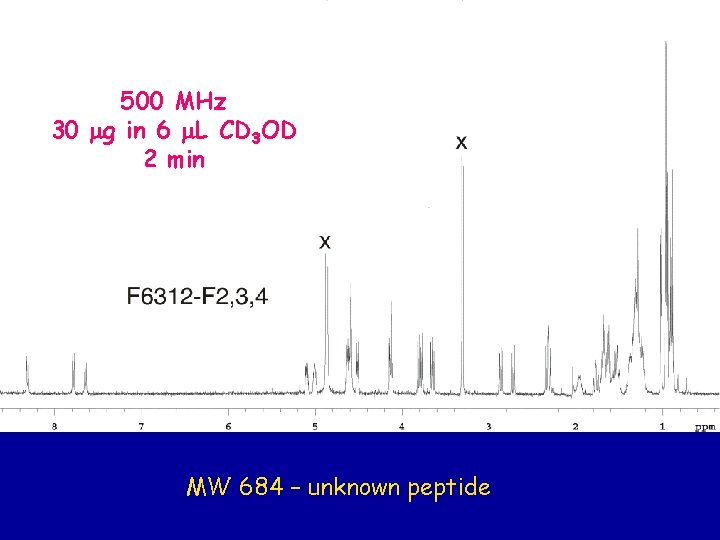
500 MHz 30 mg in 6 m. L CD 3 OD 2 min MW 684 – unknown peptide
TOCSY 40 min
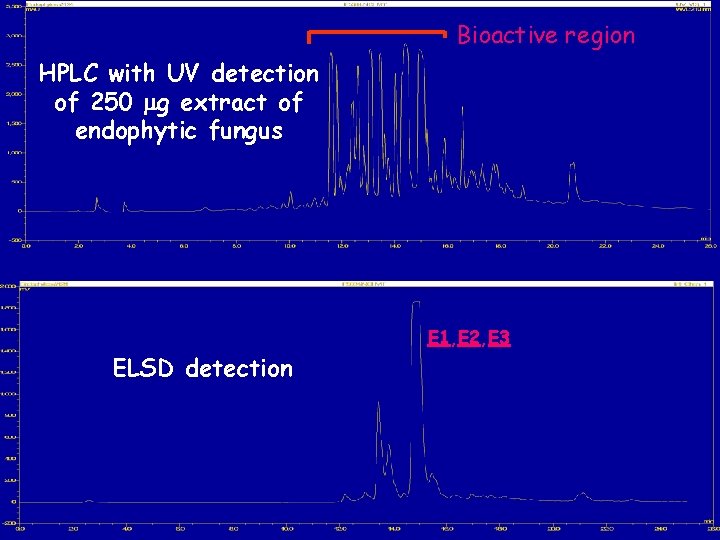
Bioactive region HPLC with UV detection of 250 mg extract of endophytic fungus E 1, E 2, E 3 ELSD detection
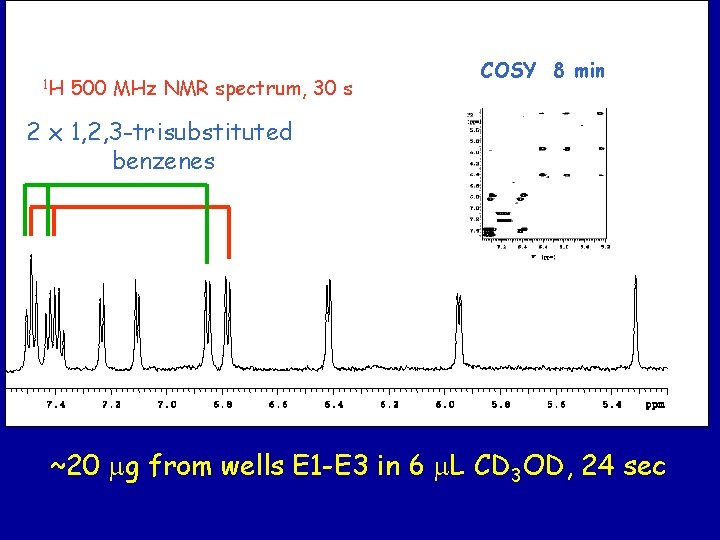
1 H 500 MHz NMR spectrum, 30 s COSY 8 min 2 x 1, 2, 3 -trisubstituted benzenes ~20 mg from wells E 1 -E 3 in 6 m. L CD 3 OD, 24 sec

Anti. Marin search with MW = 320, 0 CH 3, 2 x 1, 2, 3 -trisubstituted benzenes
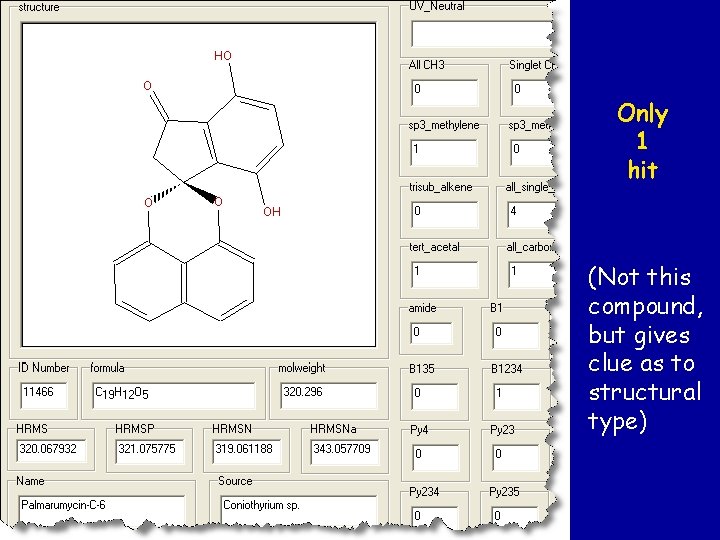
Only 1 hit (Not this compound, but gives clue as to structural type)

Other search strategies 2 x 1, 2, 3 -trisubstituted benzenes 198 hits plus 0 x CH 3 89 plus 0 x sp 3 -methylene 35 plus 1 x sp 3 -methine (none of these fit data, but all contain 1, 8 -dioxonaphthalene fragment) 3
g. HSQC 45 min g. HMBC 11 hr
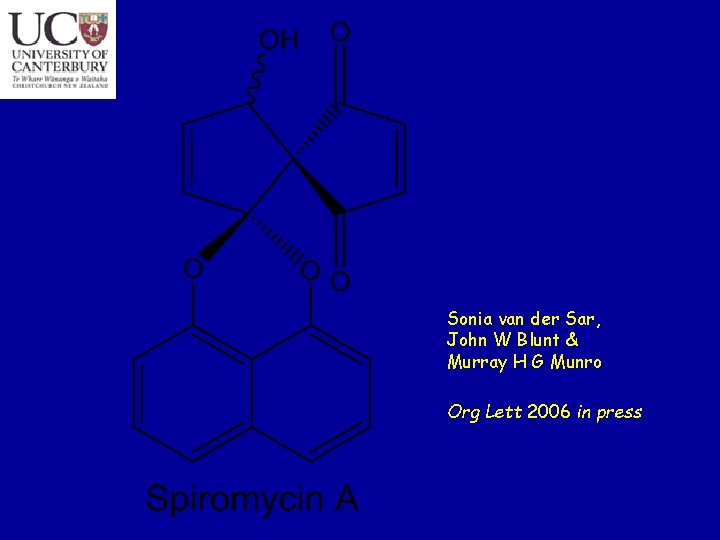
Sonia van der Sar, John W Blunt & Murray H G Munro Org Lett 2006 in press
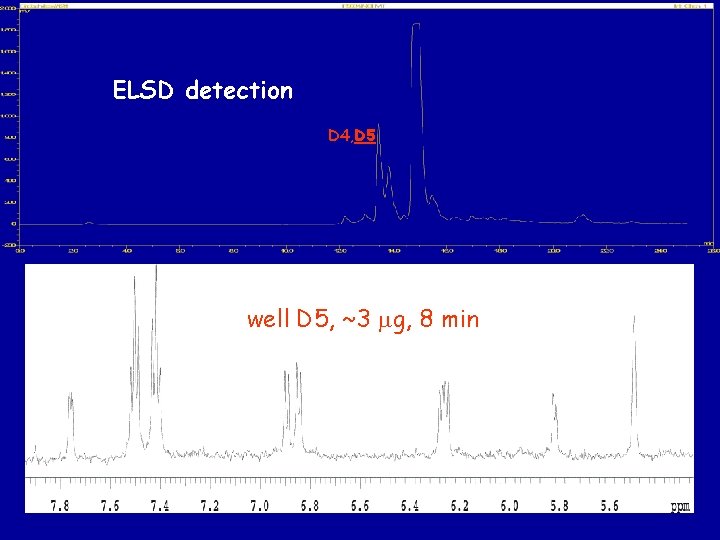
ELSD detection D 4, D 5 well D 5, ~3 mg, 8 min
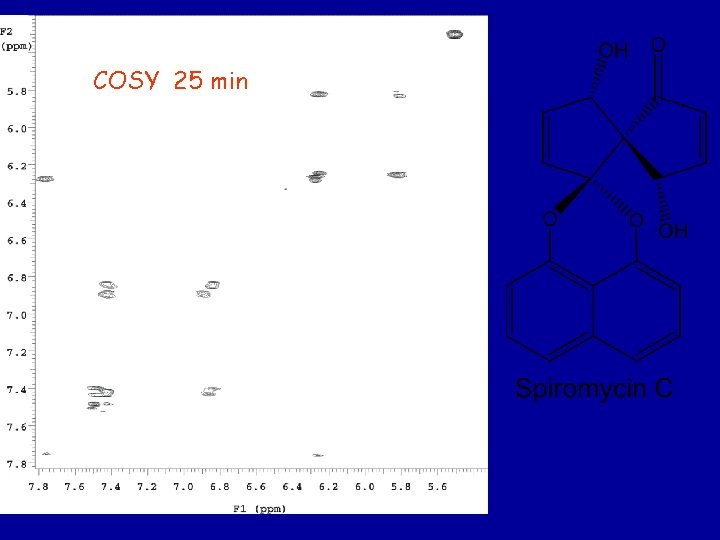
COSY 25 min
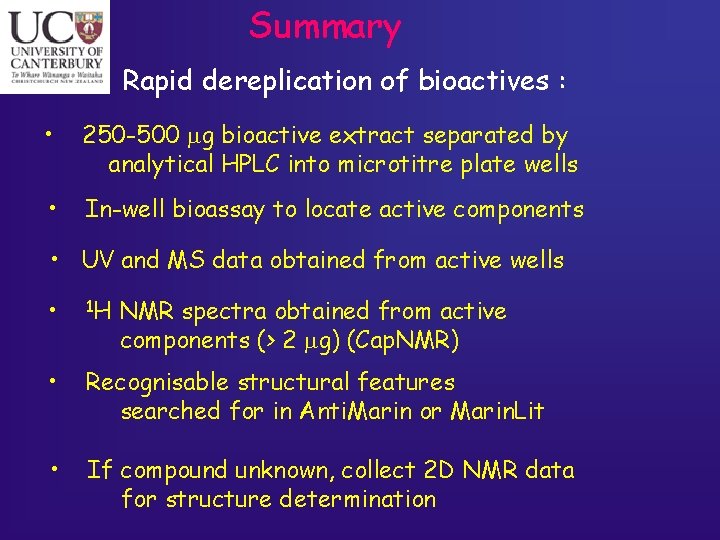
Summary Rapid dereplication of bioactives : • 250 -500 mg bioactive extract separated by analytical HPLC into microtitre plate wells • In-well bioassay to locate active components • UV and MS data obtained from active wells • 1 H • Recognisable structural features searched for in Anti. Marin or Marin. Lit • If compound unknown, collect 2 D NMR data for structure determination NMR spectra obtained from active components (> 2 mg) (Cap. NMR)

Note – all masses of samples given in this presentation are the masses of compound in the Cap. NMR microcell – these are ~65% of the amounts originally in the microtitre plate well(s) from which they were taken. The masses have been estimated from a calibration of the CHD 2 solvent peak integral in a solution of a known compound of known concentration.
- Slides: 41