Rainer Lehtonen Ph D Genomics and genetics project



- Slides: 14

Rainer Lehtonen Ph. D, Genomics and genetics project leader Metapopulation Research Group Department of Biological and Environmental Sciences, University of Helsinki Glanville fritillary butterfly genomics and genetics

Glanville fritillary butterfly – genomics and genetics �Background �Genome project �Genome assembly >> Panu Somervuo �Some NGS applications �Conclusions 2

Glanville fritillary as a model �Glanville fritillary is an internationally recognized metapopulation model system in ecological and evolutionary studies �Studied since 1991 in the Åland Islands in Finland �Data available from different populations: - Fragmented landscape vs. continuous Isolated vs. metapopulation Large vs. small Same vs. different population history Field studies, indoor & outdoor cage + laboratory experiments, controlled crosses, molecular studies 3

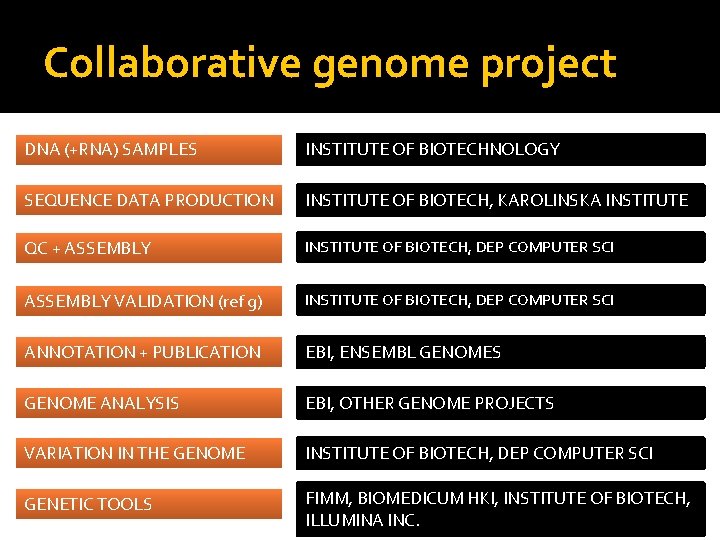
Collaborative genome project DNA (+RNA) SAMPLES INSTITUTE OF BIOTECHNOLOGY SEQUENCE DATA PRODUCTION INSTITUTE OF BIOTECH, KAROLINSKA INSTITUTE QC + ASSEMBLY INSTITUTE OF BIOTECH, DEP COMPUTER SCI ASSEMBLY VALIDATION (ref g) INSTITUTE OF BIOTECH, DEP COMPUTER SCI ANNOTATION + PUBLICATION EBI, ENSEMBL GENOMES GENOME ANALYSIS EBI, OTHER GENOME PROJECTS VARIATION IN THE GENOME INSTITUTE OF BIOTECH, DEP COMPUTER SCI GENETIC TOOLS FIMM, BIOMEDICUM HKI, INSTITUTE OF BIOTECH, ILLUMINA INC. 4

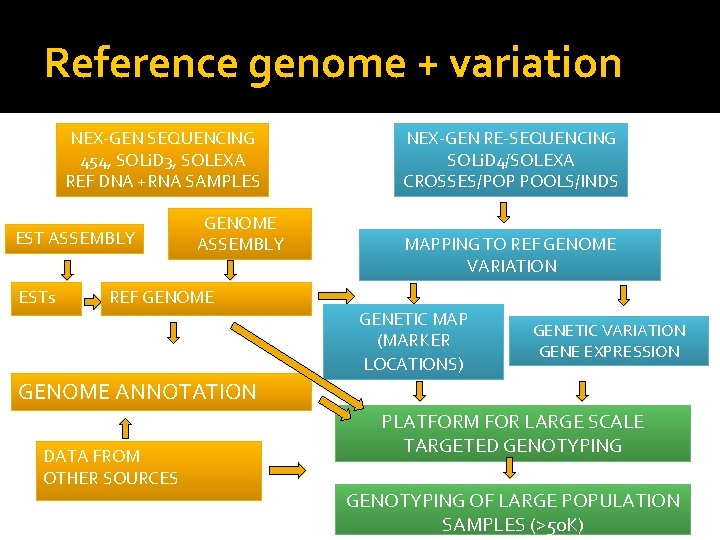
Reference genome + variation NEX-GEN SEQUENCING 454, SOLi. D 3, SOLEXA REF DNA +RNA SAMPLES EST ASSEMBLY ESTs GENOME ASSEMBLY REF GENOME NEX-GEN RE-SEQUENCING SOLi. D 4/SOLEXA CROSSES/POP POOLS/INDS MAPPING TO REF GENOME VARIATION GENETIC MAP (MARKER LOCATIONS) GENETIC VARIATION GENE EXPRESSION GENOME ANNOTATION DATA FROM OTHER SOURCES PLATFORM FOR LARGE SCALE TARGETED GENOTYPING OF LARGE POPULATION SAMPLES (>50 K)

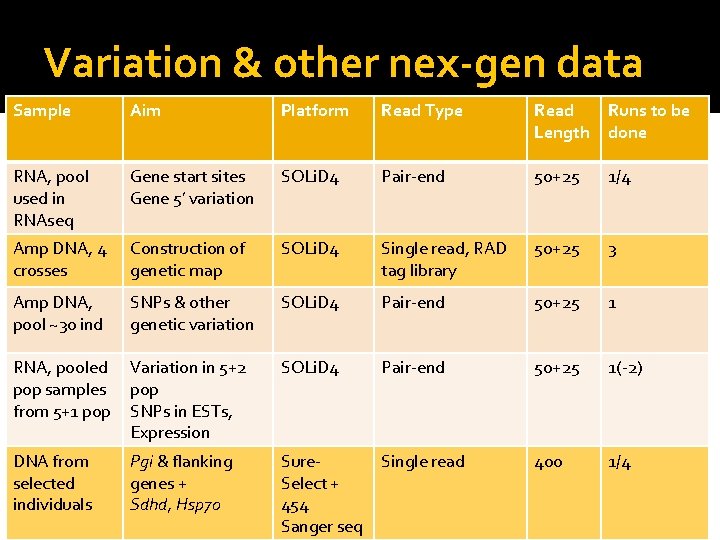
Variation & other nex-gen data Sample Aim Platform Read Type Read Length Runs to be done RNA, pool used in RNAseq Gene start sites Gene 5’ variation SOLi. D 4 Pair-end 50+25 1/4 Amp DNA, 4 crosses Construction of genetic map SOLi. D 4 Single read, RAD tag library 50+25 3 Amp DNA, pool ~30 ind SNPs & other genetic variation SOLi. D 4 Pair-end 50+25 1 RNA, pooled Variation in 5+2 pop samples pop from 5+1 pop SNPs in ESTs, Expression SOLi. D 4 Pair-end 50+25 1(-2) Single read 400 1/4 DNA from selected individuals 25. -26. 3. 2010 Pgi & flanking genes + Sdhd, Hsp 70 Sure. Select + 454 Heliconius Genome Meeting Sanger seq 6

Deep re-sequencing RAD-tag (Restriction Enzyme Associated DNA) known also as “Deep sequencing of reduced representation library” Example: Construction of a high-density genetic map: *4 controlled Spain-Finland crosses * Parents and 50 individuals from each family to be sequenced Genetic or linkage map defines an order and distance between markers based on a recombination frequency (1 c. M = 1% recombination rate) in meiosis Sure. Select (Agilent)Target Enrichment + deep sequencing with 454 Example: Population comparison of the Pgi + flanking genes (+ some other) in a sample of 24 individuals or pools 7

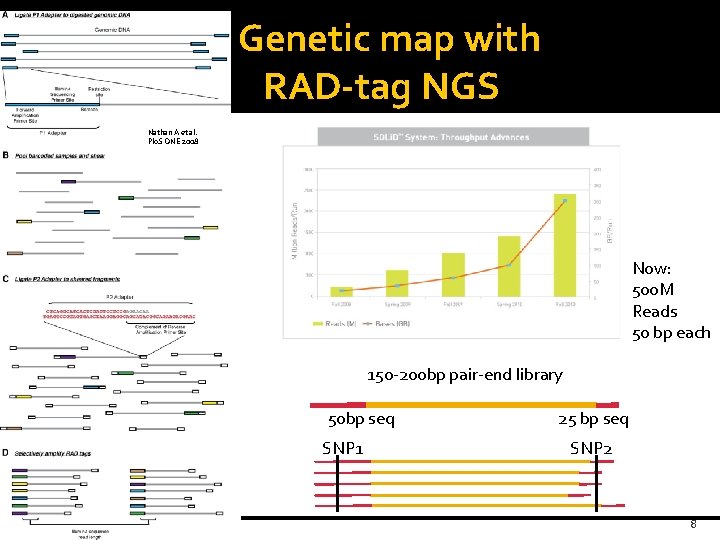
Genetic map with RAD-tag NGS Nathan A et al. Plo. S ONE 2008 Now: 500 M Reads 50 bp each 150 -200 bp pair-end library 50 bp seq SNP 1 25 bp seq SNP 2 8

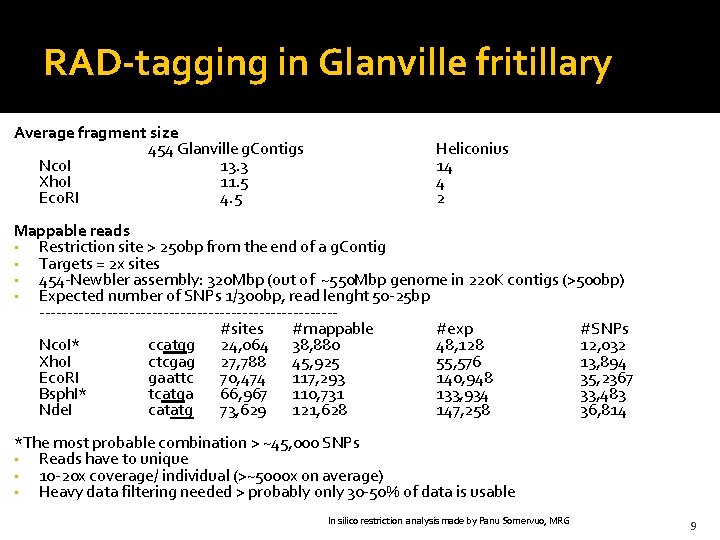
RAD-tagging in Glanville fritillary Average fragment size 454 Glanville g. Contigs Nco. I 13. 3 Xho. I 11. 5 Eco. RI 4. 5 Heliconius 14 4 2 Mappable reads • Restriction site > 250 bp from the end of a g. Contig • Targets = 2 x sites • 454 -Newbler assembly: 320 Mbp (out of ~550 Mbp genome in 220 K contigs (>500 bp) • Expected number of SNPs 1/300 bp, read lenght 50 -25 bp --------------------------- #sites #mappable #exp #SNPs Nco. I* ccatgg 24, 064 38, 880 48, 128 12, 032 Xho. I ctcgag 27, 788 45, 925 55, 576 13, 894 Eco. RI gaattc 70, 474 117, 293 140, 948 35, 2367 Bsph. I* tcatga 66, 967 110, 731 133, 934 33, 483 Nde. I catatg 73, 629 121, 628 147, 258 36, 814 *The most probable combination > ~45, 000 SNPs • Reads have to unique • 10 -20 x coverage/ individual (>~5000 x on average) • Heavy data filtering needed > probably only 30 -50% of data is usable In silico restriction analysis made by Panu Somervuo, MRG 9

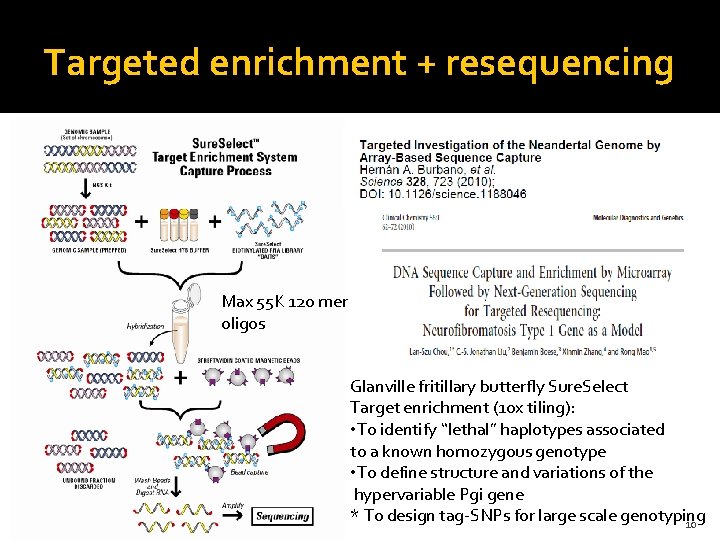
Targeted enrichment + resequencing Max 55 K 120 mer oligos Glanville fritillary butterfly Sure. Select Target enrichment (10 x tiling): • To identify “lethal” haplotypes associated to a known homozygous genotype • To define structure and variations of the hypervariable Pgi gene * To design tag-SNPs for large scale genotyping 10

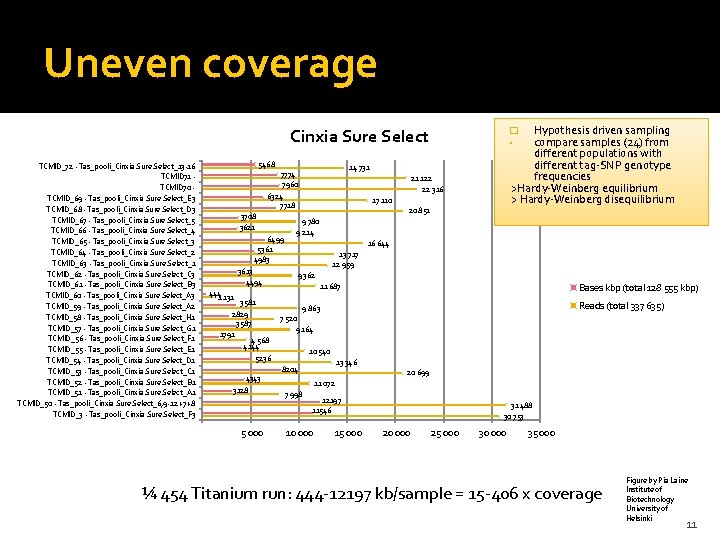
Uneven coverage Hypothesis driven sampling compare samples (24) from different populations with different tag-SNP genotype frequencies >Hardy-Weinberg equilibrium > Hardy-Weinberg disequilibrium Cinxia Sure Select 5468 TCMID_72 - Tas_pooli_Cinxia Sure Select_13 -16 TCMID 71 - TCMID 70 - TCMID_69 - Tas_pooli_Cinxia Sure Select_E 3 TCMID_68 - Tas_pooli_Cinxia Sure Select_D 3 TCMID_67 - Tas_pooli_Cinxia Sure Select_5 TCMID_66 - Tas_pooli_Cinxia Sure Select_4 TCMID_65 - Tas_pooli_Cinxia Sure Select_3 TCMID_64 - Tas_pooli_Cinxia Sure Select_2 TCMID_63 - Tas_pooli_Cinxia Sure Select_1 TCMID_62 - Tas_pooli_Cinxia Sure Select_C 3 TCMID_61 - Tas_pooli_Cinxia Sure Select_B 3 TCMID_60 - Tas_pooli_Cinxia Sure Select_A 3 TCMID_59 - Tas_pooli_Cinxia Sure Select_A 2 TCMID_58 - Tas_pooli_Cinxia Sure Select_H 1 TCMID_57 - Tas_pooli_Cinxia Sure Select_G 1 TCMID_56 - Tas_pooli_Cinxia Sure Select_F 1 TCMID_55 - Tas_pooli_Cinxia Sure Select_E 1 TCMID_54 - Tas_pooli_Cinxia Sure Select_D 1 TCMID_53 - Tas_pooli_Cinxia Sure Select_C 1 TCMID_52 - Tas_pooli_Cinxia Sure Select_B 1 TCMID_51 - Tas_pooli_Cinxia Sure Select_A 1 TCMID_50 - Tas_pooli_Cinxia Sure Select_6, 9 -12 +7+8 TCMID_3 - Tas_pooli_Cinxia Sure Select_F 3 14 731 7774 7960 6324 7718 3708 3621 6499 5361 4983 3613 4494 21 122 22 316 17 110 20 851 9 780 9 214 16 644 13 717 12 959 9 362 3128 5 000 Reads (total 337 635) 9 863 7 520 9 164 10 540 5236 4343 Bases kbp (total 128 555 kbp) 11 687 444 1 131 3581 2829 3587 1791 4 568 4144 � • 13 346 8204 20 699 11 072 7 998 12197 11546 10 000 15 000 31 488 30 753 20 000 25 000 30 000 35 000 ¼ 454 Titanium run: 444 -12197 kb/sample = 15 -406 x coverage Figure by Pia Laine Institute of Biotechnology University of Helsinki 11

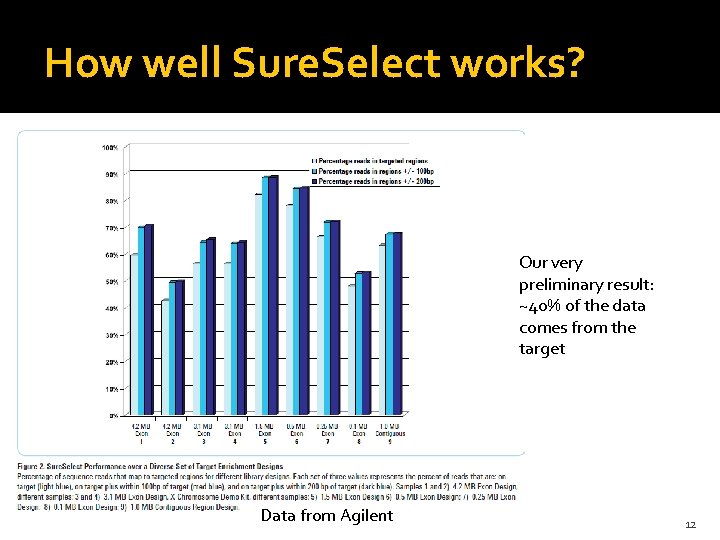
How well Sure. Select works? Our very preliminary result: ~40% of the data comes from the target Data from Agilent 12

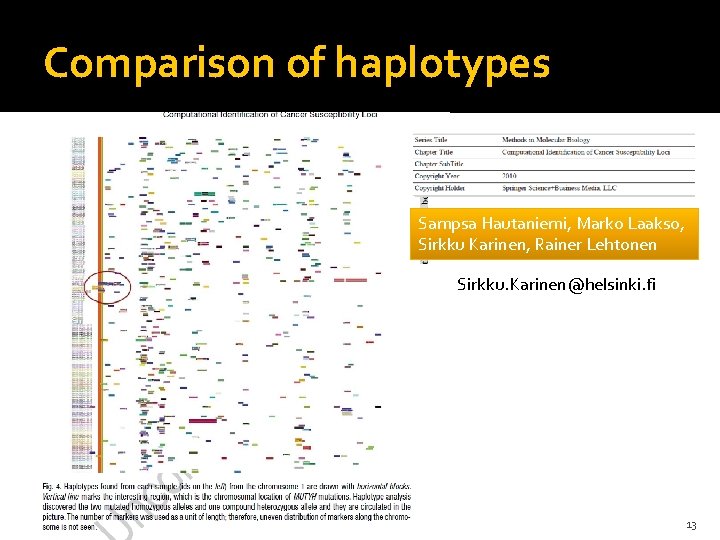
Comparison of haplotypes Sampsa Hautaniemi, Marko Laakso, Sirkku Karinen, Rainer Lehtonen Sirkku. Karinen@helsinki. fi 25. -26. 3. 2010 Heliconius Genome Meeting 13

Message �Whole genome sequencing is doable for a “non-genome” oriented research group �Most work on data filtering and analysis �Tools for data management and analysis under strong development �Down-stream efforts need to be compatible with available genome data 14