RADIATION DAMAGE TO DNA FROM NUCLEOSIDES TO THE









- Slides: 47

RADIATION DAMAGE TO DNA : FROM NUCLEOSIDES TO THE CELL Jean Cadet, Thierry Douki, Didier Gasparutto & Jean-Luc Ravanat Département de Recherche Fondamentale sur la Matière Condensée, SCIB/Laboratoire “Lésions des Acides Nucléiques”, CEA/Grenoble, France.

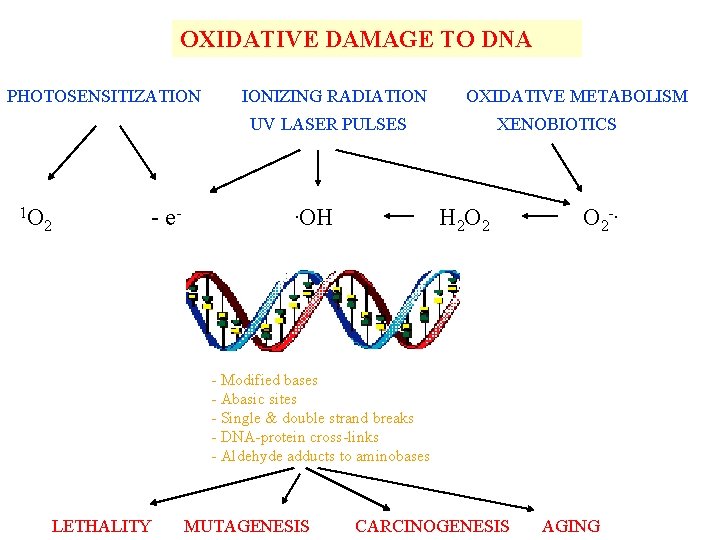
OXIDATIVE DAMAGE TO DNA PHOTOSENSITIZATION IONIZING RADIATION OXIDATIVE METABOLISM UV LASER PULSES 1 O 2 - e- . OH XENOBIOTICS H 2 O 2 O 2 -. - Modified bases - Abasic sites - Single & double strand breaks - DNA-protein cross-links - Aldehyde adducts to aminobases LETHALITY MUTAGENESIS CARCINOGENESIS AGING

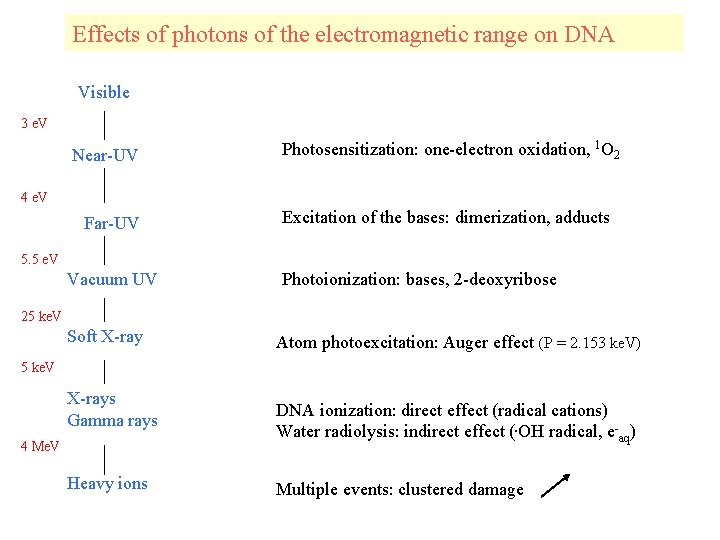
Effects of photons of the electromagnetic range on DNA Visible 3 e. V Near-UV Photosensitization: one-electron oxidation, 1 O 2 4 e. V Far-UV Excitation of the bases: dimerization, adducts 5. 5 e. V Vacuum UV Photoionization: bases, 2 -deoxyribose 25 ke. V Soft X-ray Atom photoexcitation: Auger effect (P = 2. 153 ke. V) 5 ke. V X-rays Gamma rays 4 Me. V Heavy ions DNA ionization: direct effect (radical cations) Water radiolysis: indirect effect (. OH radical, e-aq) Multiple events: clustered damage

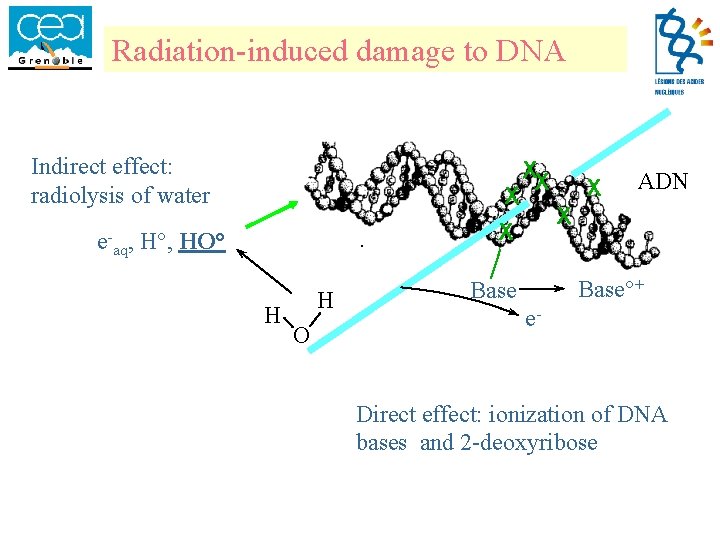
Radiation-induced damage to DNA Indirect effect: radiolysis of water X X e-aq, H°, HO° H H O XX X ADN X Base°+ Base e- Direct effect: ionization of DNA bases and 2 -deoxyribose

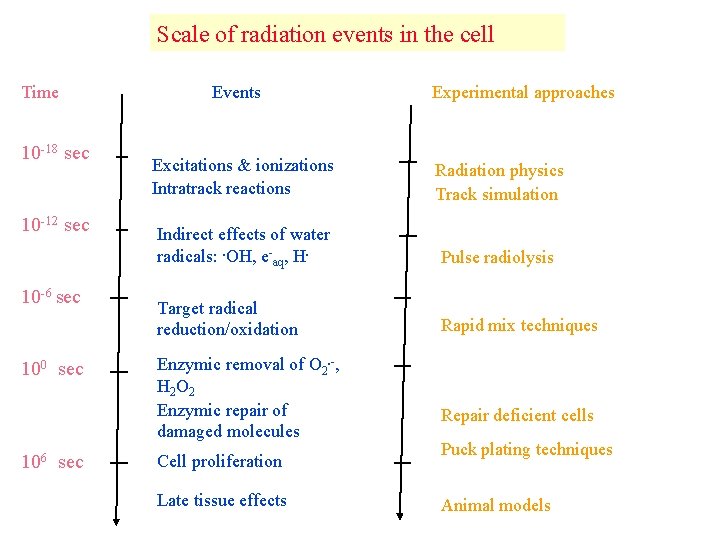
Scale of radiation events in the cell Time 10 -18 sec 10 -12 sec 10 -6 sec 100 sec 106 sec Events Experimental approaches Excitations & ionizations Intratrack reactions Radiation physics Track simulation Indirect effects of water radicals: . OH, e-aq, H. Pulse radiolysis Target radical reduction/oxidation Rapid mix techniques Enzymic removal of O 2. -, H 2 O 2 Enzymic repair of damaged molecules Cell proliferation Late tissue effects Repair deficient cells Puck plating techniques Animal models

EFFECTS OF IONIZING RADIATION ON NUCLEIC ACIDS - SINGLE LESIONS due to single events such as: . OH, ionization, secondary electrons, Auger effect …. - MULTIPLE LESIONS tandem lesions: due to either a single event (. OH or ionization ) clustered lesions due to multionization processes involving several events and/or reactive species: ionization of DNA, water radiolysis species & secondary electron

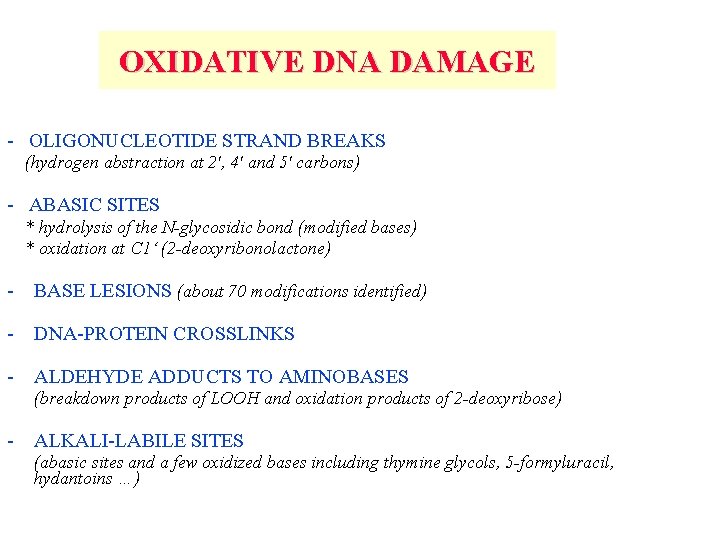
OXIDATIVE DNA DAMAGE - OLIGONUCLEOTIDE STRAND BREAKS (hydrogen abstraction at 2', 4' and 5' carbons) - ABASIC SITES * hydrolysis of the N-glycosidic bond (modified bases) * oxidation at C 1‘ (2 -deoxyribonolactone) - BASE LESIONS (about 70 modifications identified) - DNA-PROTEIN CROSSLINKS - ALDEHYDE ADDUCTS TO AMINOBASES (breakdown products of LOOH and oxidation products of 2 -deoxyribose) - ALKALI-LABILE SITES (abasic sites and a few oxidized bases including thymine glycols, 5 -formyluracil, hydantoins …)

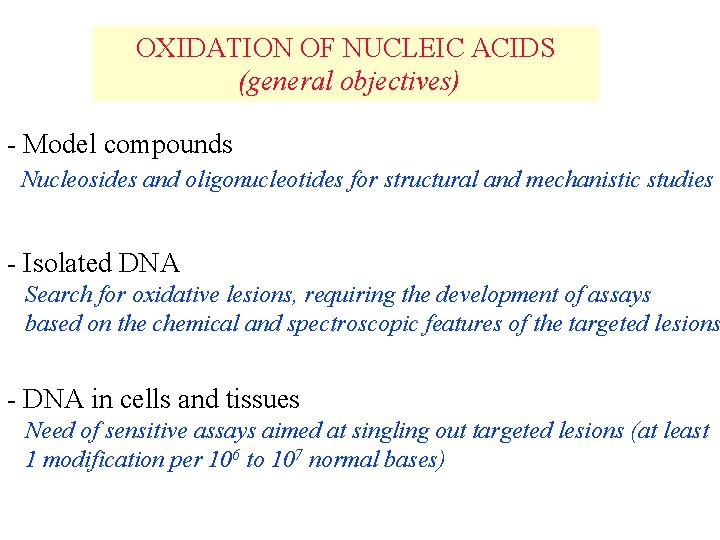
OXIDATION OF NUCLEIC ACIDS (general objectives) - Model compounds Nucleosides and oligonucleotides for structural and mechanistic studies - Isolated DNA Search for oxidative lesions, requiring the development of assays based on the chemical and spectroscopic features of the targeted lesions - DNA in cells and tissues Need of sensitive assays aimed at singling out targeted lesions (at least 1 modification per 106 to 107 normal bases)

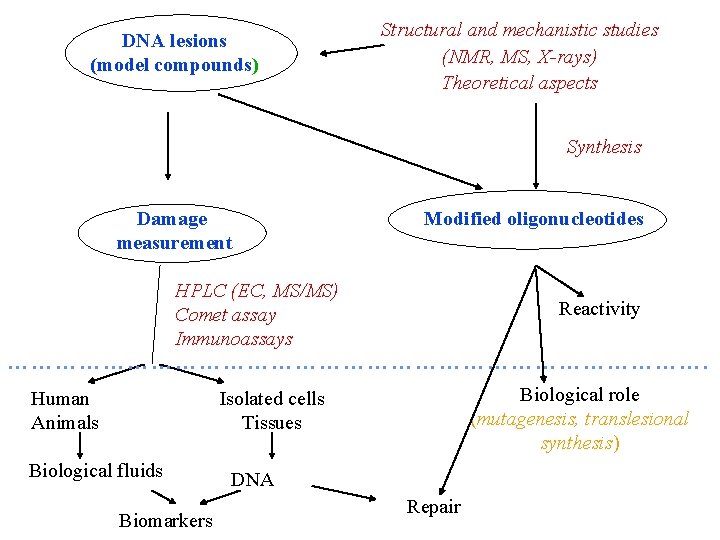
DNA lesions (model compounds) Structural and mechanistic studies (NMR, MS, X-rays) Theoretical aspects Synthesis Damage measurement Modified oligonucleotides HPLC (EC, MS/MS) Comet assay Immunoassays Reactivity ………………………………………. Human Animals Biological role (mutagenesis, translesional synthesis) Isolated cells Tissues Biological fluids Biomarkers DNA Repair

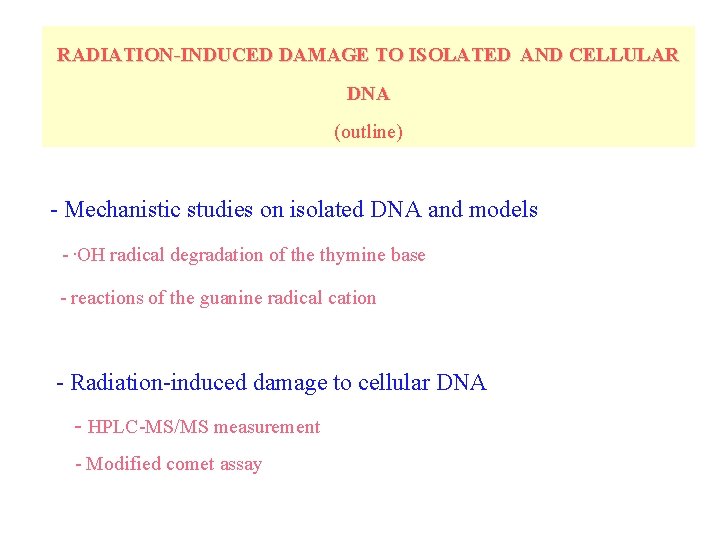
RADIATION-INDUCED DAMAGE TO ISOLATED AND CELLULAR DNA (outline) - Mechanistic studies on isolated DNA and models -. OH radical degradation of the thymine base - reactions of the guanine radical cation - Radiation-induced damage to cellular DNA - HPLC-MS/MS measurement - Modified comet assay

OXIDATIVE BASE DAMAGE TO DNA (current situation) - THYMINE: almost complete available information for isolated DNA and model compounds - CYTOSINE: comprehensive mechanism for radical oxidation of d. Cyd. Paucity of information for isolated DNA - GUANINE: complex reactions with still a strong need of further investigations on both d. Guo and isolated DNA - ADENINE: apparent lack of information (need to be further checked)

Reactions of. OH radical with thymine

OXIDATIVE DAMAGE TO GUANINE (mechanistic aspects) - Single damage: (one-electron oxidation) - Modulating effect of 8 -oxo-7, 8 -dihydroguanine (one-electron oxidation) - Tandem lesions (. OH radical, one-electron oxidation) - Charge transfer reaction within DNA

Main chemical reactions of the guanine radical cation Kasai et al, JACS, 1992 Cadet et al, JACS 1994 Gasparutto et al, JACS 1998 Ravanat et al, JACS 2003

RADICAL OXIDATION OF GUANINE AND ADENINE IN ISOLATED DNA (similarities and differences) - Similar degradation products from 8 -hydroxy-7, 8 -dihydro purinyl radicals: 8 -oxo-7, 8 -dihydro- and Fapy purine derivatives - 2 -Hydroxyadenine: barely detectable with yield much lower than that of Fapy. Ade - On the overall degradation of guanine 10 -fold less efficient than that of adenine in isolated DNA but not in nucleosides. - Unknown reasons of the apparent lower susceptibility of adenine to both. OH radical and one-electron oxidants.

Modulating effect of 8 -oxod. Guo on one-electron oxidative damage to DNA - Ionization potential of 8 -oxod. Guo is lower than that of other DNA nucleosides including d. Guo (Prat et al, J. Am. Chem. Soc. , 120, 845 -846, 1998) - Rate constant for one-electron oxidation of 8 -oxod. Guo is about 2 orders of magnitude higher with respect to that of d. Guo (Steenken et al, J. Am. Chem. Soc. , 122, 2373 -2374, 2000) - 8 -Oxod. Guo could be the ultimate sink of one-electron oxidation process within DNA

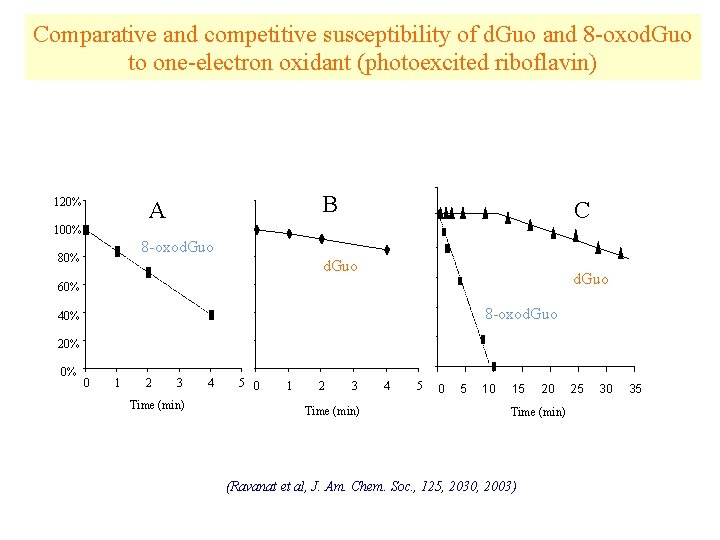
Comparative and competitive susceptibility of d. Guo and 8 -oxod. Guo to one-electron oxidant (photoexcited riboflavin) 120% B A 100% C 8 -oxod. Guo 80% d. Guo 60% 8 -oxod. Guo 40% 20% 0% 0 1 2 3 Time (min) 4 5 0 5 10 15 20 Time (min) (Ravanat et al, J. Am. Chem. Soc. , 125, 2030, 2003) 25 30 35

Radical oxidation reactions of isolated 2’-deoxyguanosine

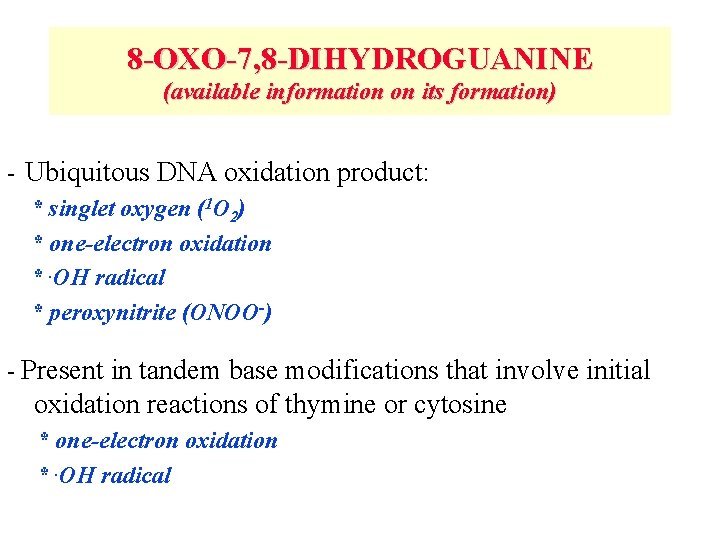
8 -OXO-7, 8 -DIHYDROGUANINE (available information on its formation) - Ubiquitous DNA oxidation product: * singlet oxygen (1 O 2) * one-electron oxidation *. OH radical * peroxynitrite (ONOO-) - Present in tandem base modifications that involve initial oxidation reactions of thymine or cytosine * one-electron oxidation *. OH radical

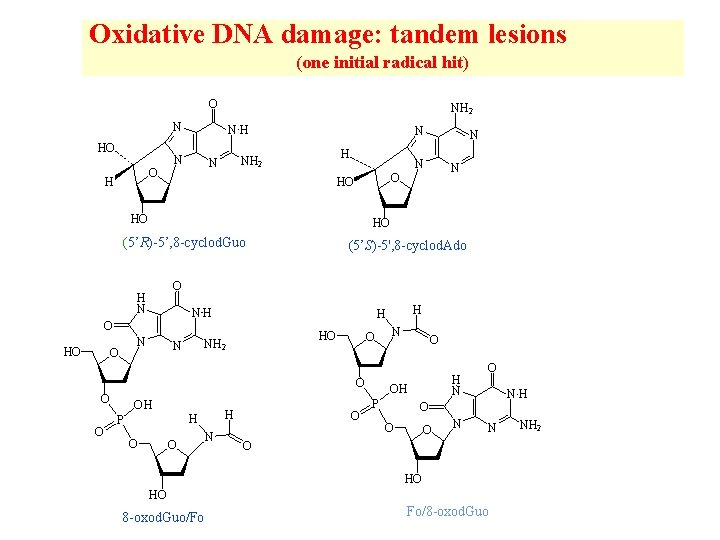
Oxidative DNA damage: tandem lesions (one initial radical hit) O NH 2 N HO O H NH N N H NH 2 N HO (5’S)-5', 8 -cyclod. Ado O H N NH O N O HO NH 2 N O OH H H P O O N H H O O O N HO (5’R)-5’, 8 -cyclod. Guo HO N O N O H N OH P O NH O O O N N O HO HO 8 -oxod. Guo/Fo Fo/8 -oxod. Guo NH 2

Mechanism of formation of 8 -oxod. Guo/formamido tandem lesions (Douki et al, Chem. Res. Toxicol. , 2002)

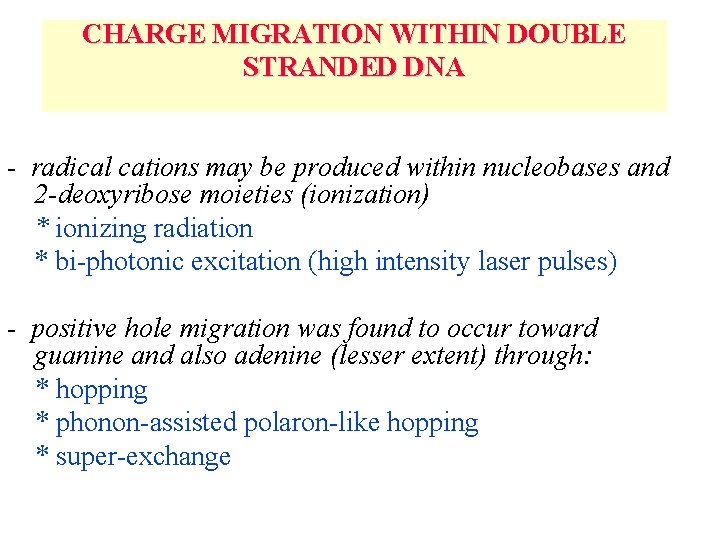
CHARGE MIGRATION WITHIN DOUBLE STRANDED DNA - radical cations may be produced within nucleobases and 2 -deoxyribose moieties (ionization) * ionizing radiation * bi-photonic excitation (high intensity laser pulses) - positive hole migration was found to occur toward guanine and also adenine (lesser extent) through: * hopping * phonon-assisted polaron-like hopping * super-exchange

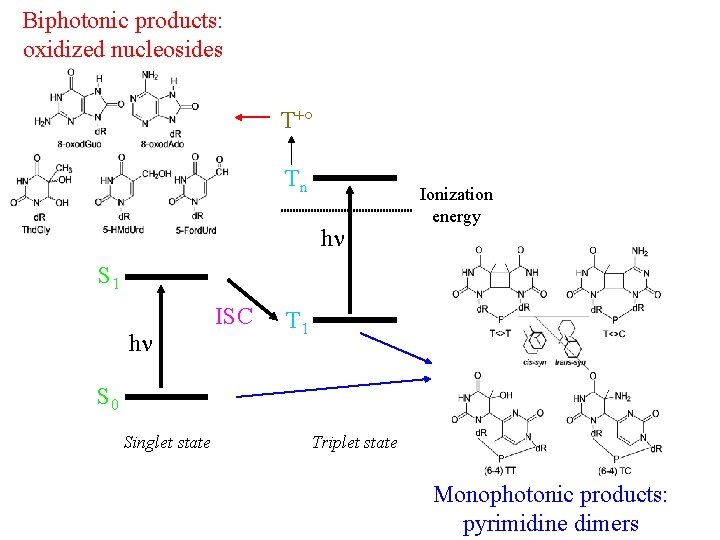
Biphotonic products: oxidized nucleosides T +° Tn hn Ionization energy S 1 ISC hn T 1 S 0 Singlet state Triplet state Monophotonic products: pyrimidine dimers

Effects of the intensity on the quantum yield of formation of oxidized nucleosides upon exposure of DNA to UV laser pulses.

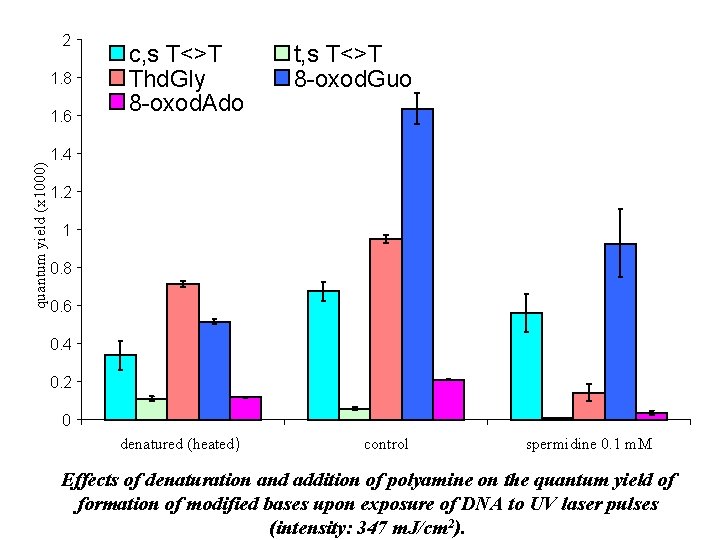
2 1. 8 quantum yield (x 1000) 1. 6 c, s T<>T Thd. Gly 8 -oxod. Ado t, s T<>T 8 -oxod. Guo denatured (heated) control 1. 4 1. 2 1 0. 8 0. 6 0. 4 0. 2 0 spermidine 0. 1 m. M Effects of denaturation and addition of polyamine on the quantum yield of formation of modified bases upon exposure of DNA to UV laser pulses (intensity: 347 m. J/cm 2).

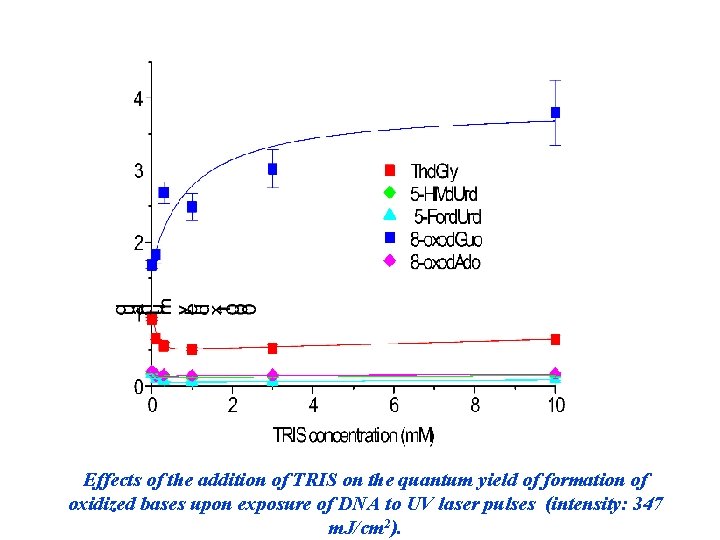
Effects of the addition of TRIS on the quantum yield of formation of oxidized bases upon exposure of DNA to UV laser pulses (intensity: 347 m. J/cm 2).

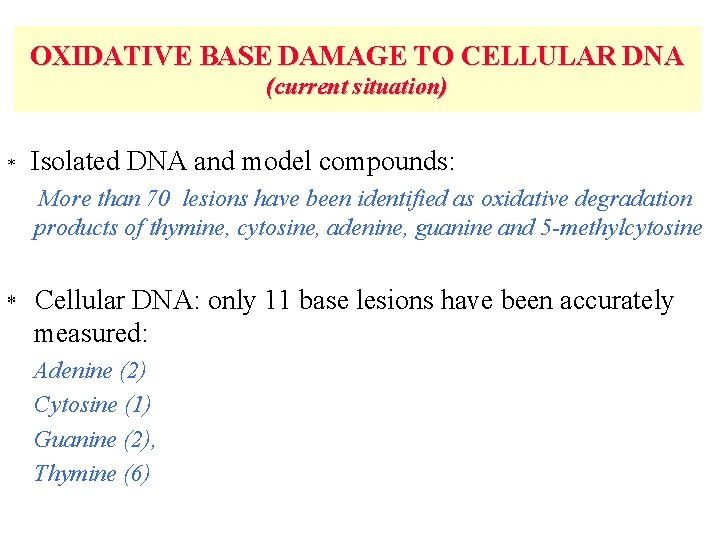
OXIDATIVE BASE DAMAGE TO CELLULAR DNA (current situation) * Isolated DNA and model compounds: More than 70 lesions have been identified as oxidative degradation products of thymine, cytosine, adenine, guanine and 5 -methylcytosine * Cellular DNA: only 11 base lesions have been accurately measured: Adenine (2) Cytosine (1) Guanine (2), Thymine (6)

MEASUREMENT OF OXIDATIVE BASE DAMAGE TO DNA - As DNA fragments (bases, nucleosides or nucleotides) - In whole DNA - In intact cells

MEASUREMENT OF OXIDATIVE BASE DAMAGE TO CELLULAR DNA (individual measurement) - Chromatographic methods: HPLC-MS/MS Optimization of DNA extraction conditions - Applications: Effects of γ-rays and heavy ions on human monocytes

DNA extraction for chromatographic assays * Still a critical step * Various methods have been proposed with or without phenol The background level is within the range of one up to several 8 -oxod. Guo residue per 106 bp * Chaotropic method associated with a metal chelator (desferioxamine) The background level is lower than one 8 -oxod. Guo residue per 106 bp (Helbock et al, PNAS, 95, 288 -93, 1998; Ravanat et al, Carcinogenesis, 23, 1911 -8, 2002)

OXIDATIVE DNA BASE DAMAGE HPLC-MS/MS (electrospray ionization mode) measurements - Most recent method (tandem mass spectrometers) - More sensitive than any other chromatographic methods by about a factor of 10 (this depends on the targeted lesion) - More straightforward than GC-MS (no derivatization) and versatile than HPLC-ECD (almost all compounds can be detected) - Extension to more sensitive analytical methods (micro-HPLC, capillary electrophoresis)

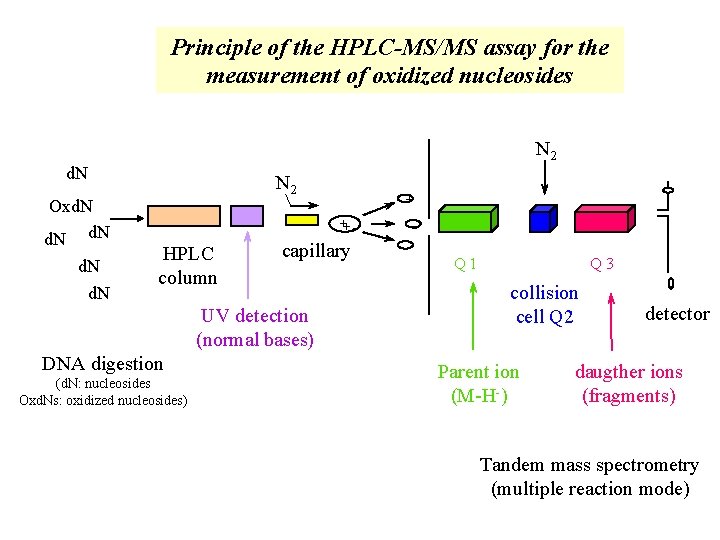
Principle of the HPLC-MS/MS assay for the measurement of oxidized nucleosides N 2 d. N Oxd. N d. N N 2 + ++ - HPLC column capillary UV detection (normal bases) DNA digestion (d. N: nucleosides Oxd. Ns: oxidized nucleosides) - Q 1 Q 3 collision cell Q 2 Parent ion (M-H-) detector daugther ions (fragments) Tandem mass spectrometry (multiple reaction mode)

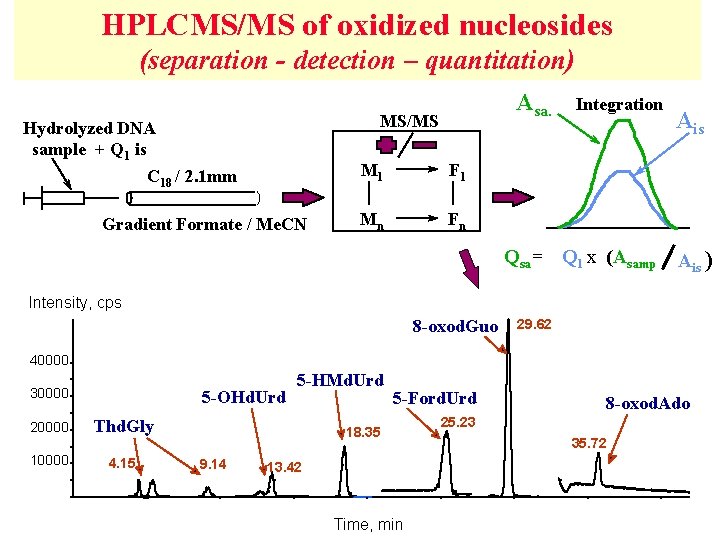
HPLCMS/MS of oxidized nucleosides (separation - detection – quantitation) Asa. MS/MS Hydrolyzed DNA sample + Q 1 is C 18 / 2. 1 mm Gradient Formate / Me. CN M 1 F 1 Mn Fn Integration Qsa= Q 1 x (Asamp Ais ) Intensity, cps 8 -oxod. Guo 29. 62 40000 30000 20000 10000 5 -OHd. Urd 5 -HMd. Urd Thd. Gly 4. 15 5 -Ford. Urd 18. 35 9. 14 13. 42 Time, min 8 -oxod. Ado 25. 23 35. 72

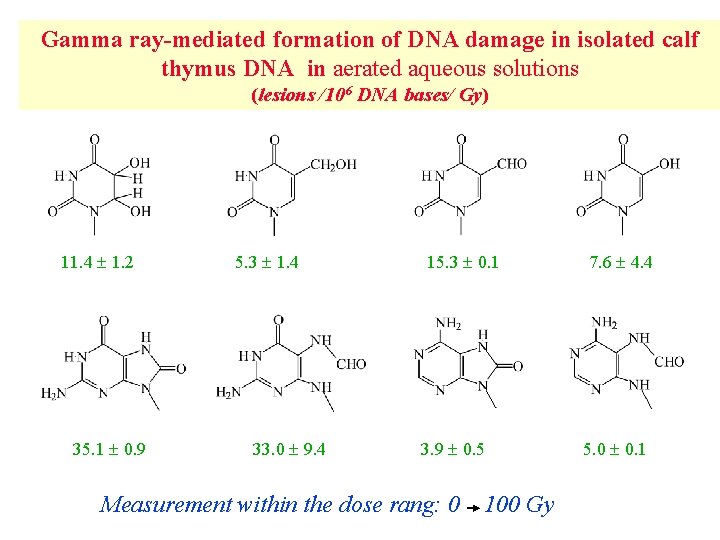
Gamma ray-mediated formation of DNA damage in isolated calf thymus DNA in aerated aqueous solutions (lesions /106 DNA bases/ Gy) 11. 4 1. 2 35. 1 0. 9 5. 3 1. 4 33. 0 9. 4 15. 3 0. 1 3. 9 0. 5 Measurement within the dose rang: 0 100 Gy 7. 6 4. 4 5. 0 0. 1

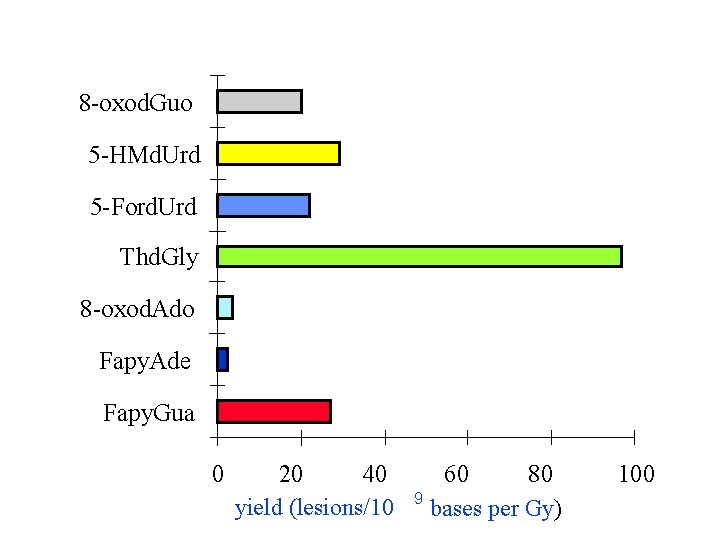
8 -oxod. Guo 5 -HMd. Urd 5 -Ford. Urd Thd. Gly 8 -oxod. Ado Fapy. Ade Fapy. Gua 0 20 40 yield (lesions/10 60 80 9 bases per Gy) 100

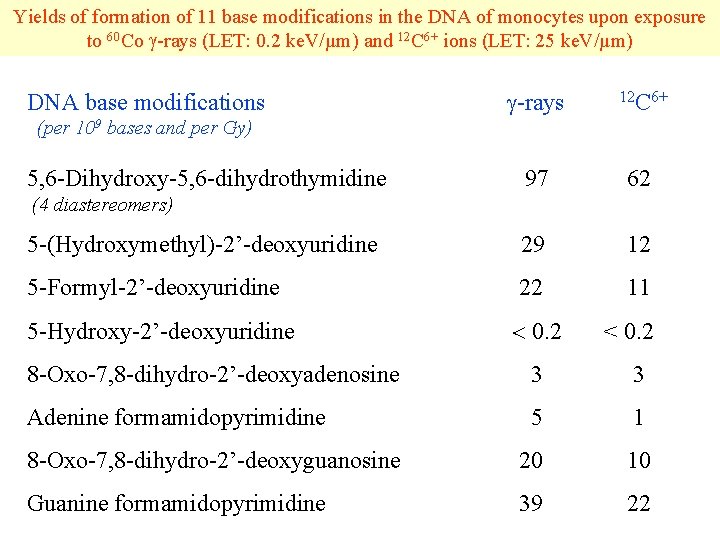
Yields of formation of 11 base modifications in the DNA of monocytes upon exposure to 60 Co -rays (LET: 0. 2 ke. V/µm) and 12 C 6+ ions (LET: 25 ke. V/µm) -rays 12 C 6+ 97 62 5 -(Hydroxymethyl)-2’-deoxyuridine 29 12 5 -Formyl-2’-deoxyuridine 22 11 5 -Hydroxy-2’-deoxyuridine 0. 2 DNA base modifications (per 109 bases and per Gy) 5, 6 -Dihydroxy-5, 6 -dihydrothymidine (4 diastereomers) < 0. 2 8 -Oxo-7, 8 -dihydro-2’-deoxyadenosine 3 3 Adenine formamidopyrimidine 5 1 8 -Oxo-7, 8 -dihydro-2’-deoxyguanosine 20 10 Guanine formamidopyrimidine 39 22

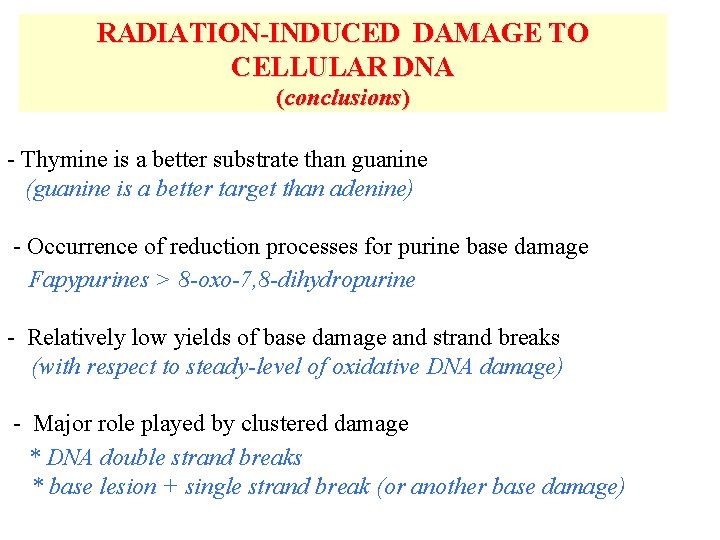
RADIATION-INDUCED DAMAGE TO CELLULAR DNA (conclusions) - Thymine is a better substrate than guanine (guanine is a better target than adenine) - Occurrence of reduction processes for purine base damage Fapypurines > 8 -oxo-7, 8 -dihydropurine - Relatively low yields of base damage and strand breaks (with respect to steady-level of oxidative DNA damage) - Major role played by clustered damage * DNA double strand breaks * base lesion + single strand break (or another base damage)

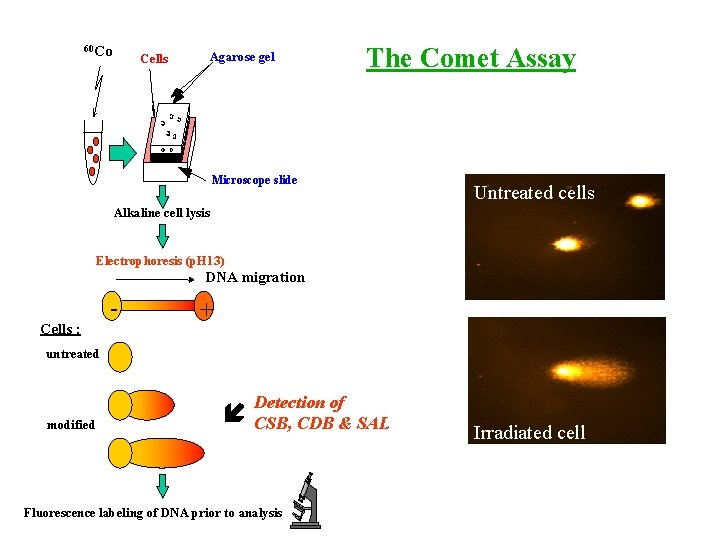
MEASUREMENT OF OXIDATIVE BASE DAMAGE TO CELLULAR DNA - Isolated cells: Modified comet assay (use of DNA repair enzymes to convert base damage into strand breaks) More sensitive but less specific than HPLC-MS/MS - Applications: Effects of ionizing radiation

60 Co Cells Agarose gel The Comet Assay Microscope slide Untreated cells Alkaline cell lysis Electrophoresis (p. H 13) DNA migration Cells : - + untreated Detection of modified í CSB, CDB & SAL Fluorescence labeling of DNA prior to analysis Irradiated cell

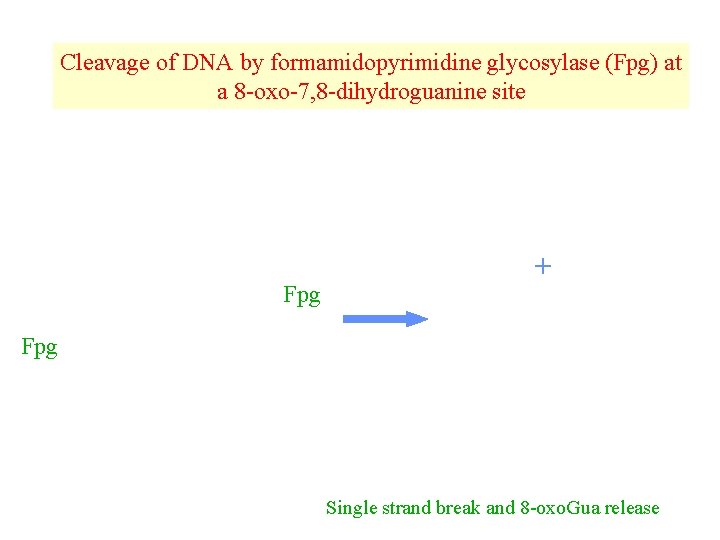
Cleavage of DNA by formamidopyrimidine glycosylase (Fpg) at a 8 -oxo-7, 8 -dihydroguanine site Fpg + Fpg Single strand break and 8 -oxo. Gua release

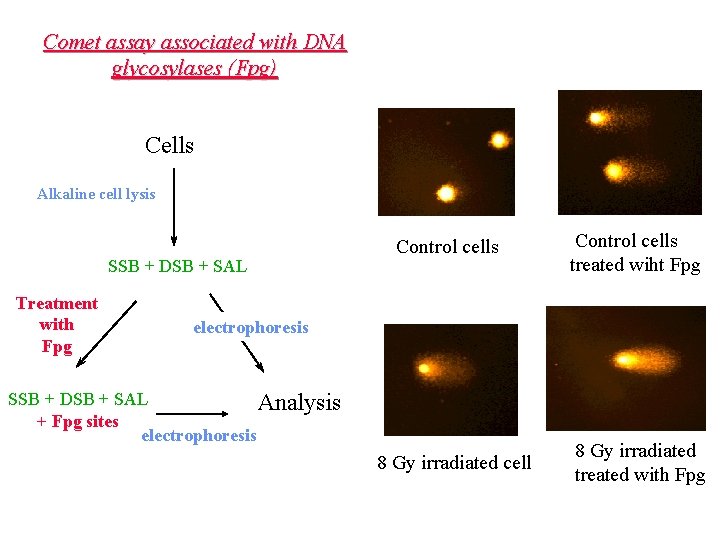
Comet assay associated with DNA glycosylases (Fpg) Cells Alkaline cell lysis SSB + DSB + SAL Treatment with Fpg Control cells treated wiht Fpg electrophoresis SSB + DSB + SAL Analysis + Fpg sites electrophoresis 8 Gy irradiated cell 8 Gy irradiated treated with Fpg

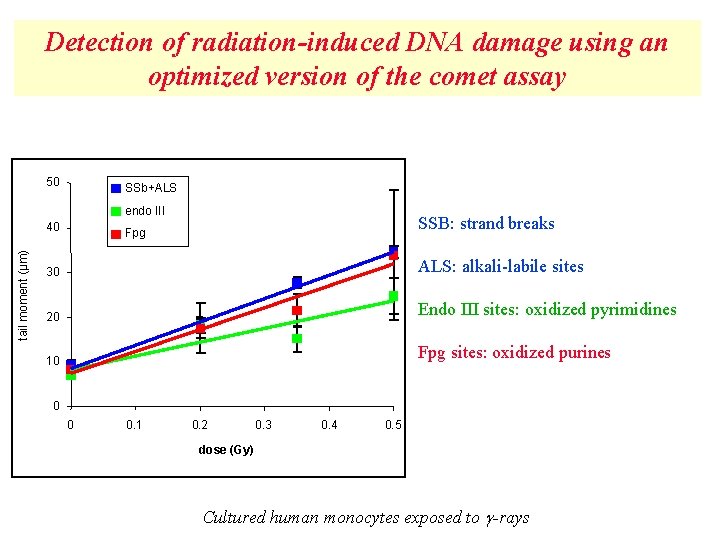
Detection of radiation-induced DNA damage using an optimized version of the comet assay 50 SSb+ALS endo III tail moment (µm) 40 SSB: strand breaks Fpg 30 ALS: alkali-labile sites 20 Endo III sites: oxidized pyrimidines 10 Fpg sites: oxidized purines 0 0 0. 1 0. 2 0. 3 0. 4 0. 5 dose (Gy) Cultured human monocytes exposed to -rays

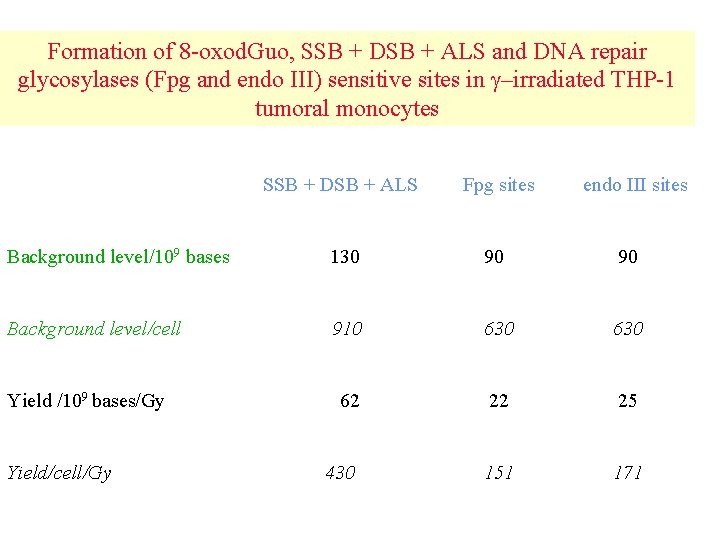
Formation of 8 -oxod. Guo, SSB + DSB + ALS and DNA repair glycosylases (Fpg and endo III) sensitive sites in –irradiated THP-1 tumoral monocytes SSB + DSB + ALS Fpg sites Background level/109 bases 130 90 90 Background level/cell 910 630 62 22 25 430 151 171 Yield /109 bases/Gy Yield/cell/Gy endo III sites

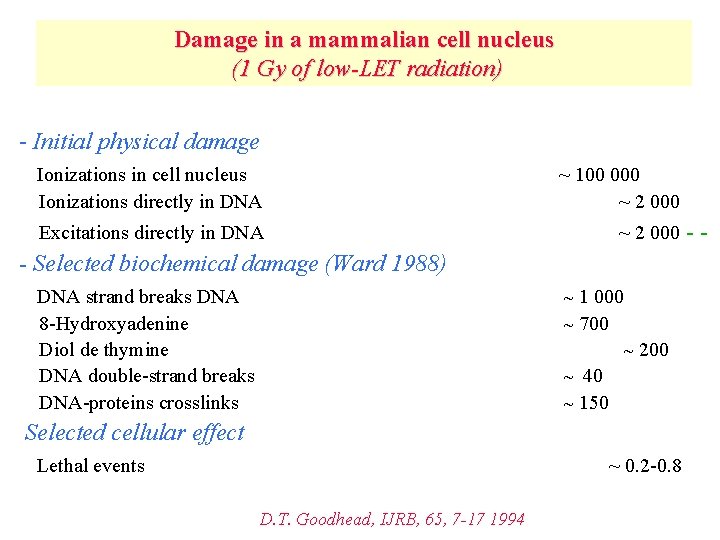
Damage in a mammalian cell nucleus (1 Gy of low-LET radiation) - Initial physical damage Ionizations in cell nucleus Ionizations directly in DNA ~ 100 000 ~ 2 000 Excitations directly in DNA ~ 2 000 - Selected biochemical damage (Ward 1988) DNA strand breaks DNA 8 -Hydroxyadenine Diol de thymine DNA double-strand breaks DNA-proteins crosslinks ~ 1 000 ~ 700 ~ 200 ~ 40 ~ 150 Selected cellular effect Lethal events ~ 0. 2 -0. 8 D. T. Goodhead, IJRB, 65, 7 -17 1994 --

RADIATON DAMAGE TO DNA (future work) - Clustered damage - DNA-protein crosslinks - DNA-aldehyde adducts (LOOH) - Bystander effects - Effects of electrons of low energy

OXIDATIVE DAMAGE TO CELLULAR DNA (conclusion) - The steady-state level of the main oxidized bases is within the range of 1 lesion per 106 - 107 normal bases. - The level of radical induced 8 -oxo. Gua is lower than that of Fapy. Gua and thymine glycol. - The level of 8 -oxo. Ade and Fapy. Ade is about 10 -fold lower than that of 8 -oxo. Gua. - 2 -Hydroxyadenine is not detectable.

OXIDATIVE DAMAGE TO CELLULAR DNA (ESCODD) - “European Standards Committee on Oxidative DNA Damage”: set in 1997 with EC funding over the period 2000 -2003; it has involved 25 member laboratories in Europe and one in Japan - Objectives: to standardize and validate procedures for measuring 8 -oxod. Guo as a biomarker of DNA oxidation - Levels of 8 -oxod. Guo (0. 5) and Fpg-sensitive sites (0. 1) per 106 bases in the DNA of human lymphocytes by HPLC and enzymic methods - It will be necessary to re-examine anti-oxidant studies that are based on claims of 8 oxod. Guo higher values than 1 per 106 bases (Collins et al, Arch. Biochem. Biophys. , 423, 57 -65, 2004)