Quantitative Protein Analysis of CYP 450 Induction via



- Slides: 23

Quantitative Protein Analysis of CYP 450 Induction via LC-MRM Analysis

Cytochrome P 450 Proteins − Cytochrome P 450 enzymes are mainly expressed in liver and are responsible for oxidative metabolism of drugs, environmental pollutants, carcinogens, etc − Cytochrome P 450 Family of Enzymes – 70 p 450 protein families in humans – Over 200 different subfamilies / isoforms – Each isoform has different substrate specificities, varied inducibility by different drugs − Important in drug development – Changes in expression of specific isoforms provide information on toxicity of different drugs – Individual patient basal expression levels affect responsiveness to drugs 2 © 2010 AB SCIEX

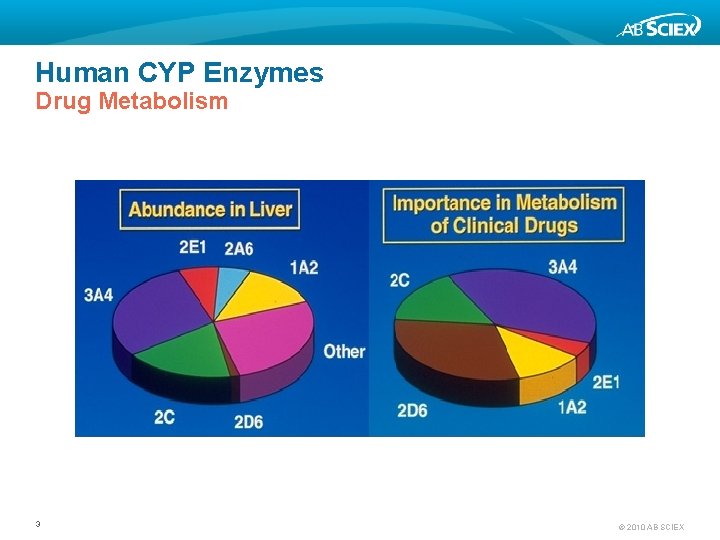
Human CYP Enzymes Drug Metabolism 3 © 2010 AB SCIEX

Current Methodologies for Assessing CYP 450 Induction − m. RNA techniques – Measure the amount of messenger RNA expressed for each enzyme isoform – Assesses only CYP induction from gene transcription changes − Enzymatic activity – Induction by quantifying the metabolite of a CYP-specific probe substrate generated from treated hepatocytes – Relies on the specificity of enzymatic conversion of probe substrates, and a different probe substrate is required for each CYP isozyme which is not always possible − Western Blotting techniques – Measures the actual protein levels of CYP 450 enzymes using isoform-specific antibodies. Currently there are only a few good antibodies that are isoform specific Major Challenges in Induction Assay: Assay selectivity, and sensitivity; Different technical expertise and equipment are needed for m. RNA or Western Blotting assessment 4 © 2010 AB SCIEX

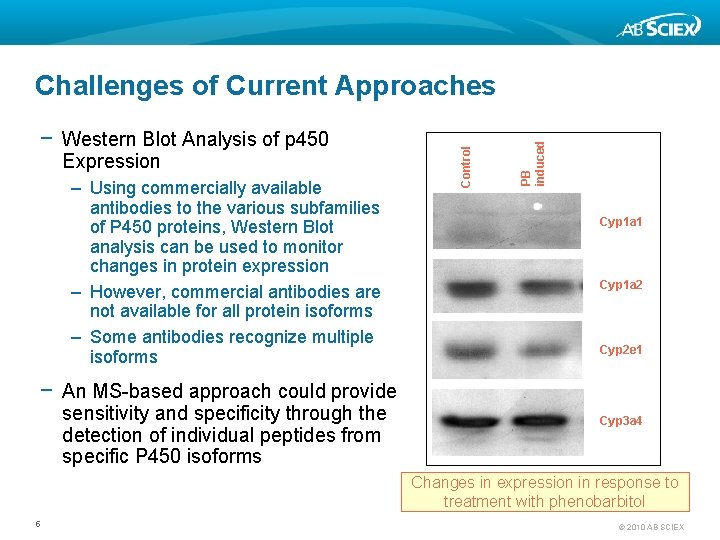
Expression – Using commercially available antibodies to the various subfamilies of P 450 proteins, Western Blot analysis can be used to monitor changes in protein expression – However, commercial antibodies are not available for all protein isoforms – Some antibodies recognize multiple isoforms PB induced − Western Blot Analysis of p 450 Control Challenges of Current Approaches Cyp 1 a 1 Cyp 1 a 2 Cyp 2 e 1 − An MS-based approach could provide sensitivity and specificity through the detection of individual peptides from specific P 450 isoforms Cyp 3 a 4 Changes in expression in response to treatment with phenobarbitol 5 © 2010 AB SCIEX

CYP Induction Assay: LC-MS/MS Solution − An LC-MS/MS-based approach can provide sensitivity and specificity through the detection of individual peptides from specific CYP 450 isoforms − A fast MS scan speed and the Scheduled MRM™ Algorithm allows for multiplexed protein quantitation − A CYP 450 protein assay kit including all reagents, sample preparation procedure and established LC/MS/MS conditions provides easy protein quantitation for human induction studies using current DMPK resources 6 © 2010 AB SCIEX

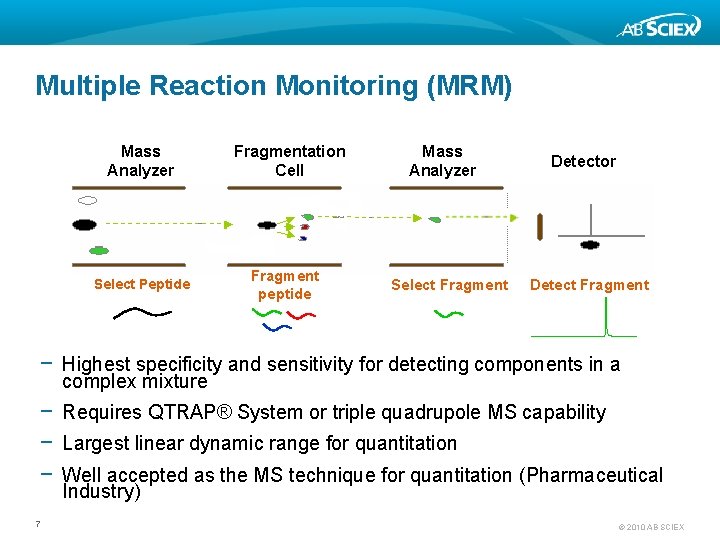
Multiple Reaction Monitoring (MRM) Mass Analyzer Select Peptide Fragmentation Cell Fragment peptide Mass Analyzer Select Fragment Detector Detect Fragment − Highest specificity and sensitivity for detecting components in a complex mixture − Requires QTRAP® System or triple quadrupole MS capability − Largest linear dynamic range for quantitation − Well accepted as the MS technique for quantitation (Pharmaceutical Industry) 7 © 2010 AB SCIEX

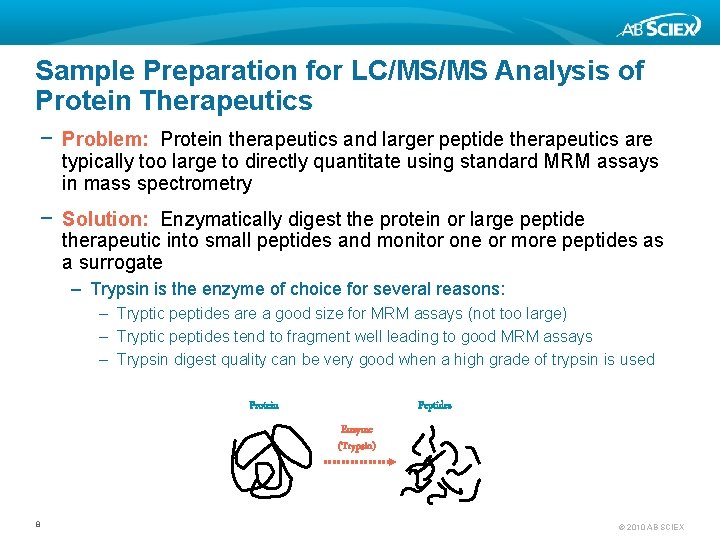
Sample Preparation for LC/MS/MS Analysis of Protein Therapeutics − Problem: Protein therapeutics and larger peptide therapeutics are typically too large to directly quantitate using standard MRM assays in mass spectrometry − Solution: Enzymatically digest the protein or large peptide therapeutic into small peptides and monitor one or more peptides as a surrogate – Trypsin is the enzyme of choice for several reasons: – Tryptic peptides are a good size for MRM assays (not too large) – Tryptic peptides tend to fragment well leading to good MRM assays – Trypsin digest quality can be very good when a high grade of trypsin is used Protein Peptides Enzyme (Trypsin) 8 © 2010 AB SCIEX

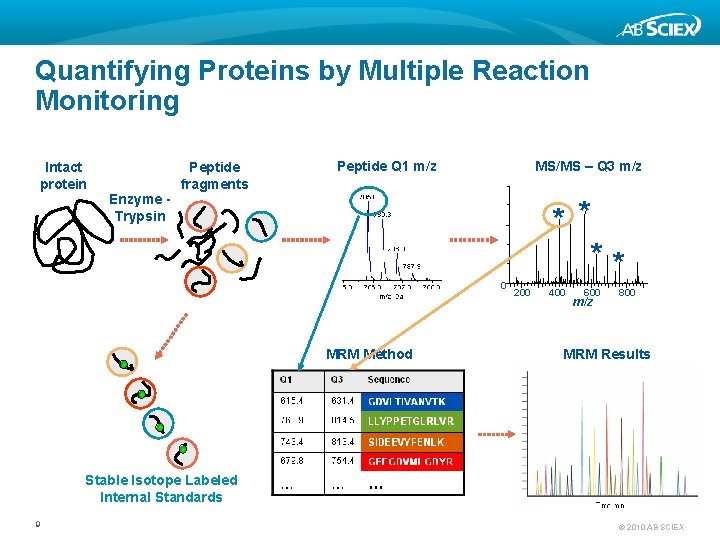
Quantifying Proteins by Multiple Reaction Monitoring Intact protein Enzyme Trypsin Peptide fragments Peptide Q 1 m/z MS/MS – Q 3 m/z 0 MRM Method * * ** 200 400 600 m/z 800 MRM Results Stable Isotope Labeled Internal Standards 9 © 2010 AB SCIEX

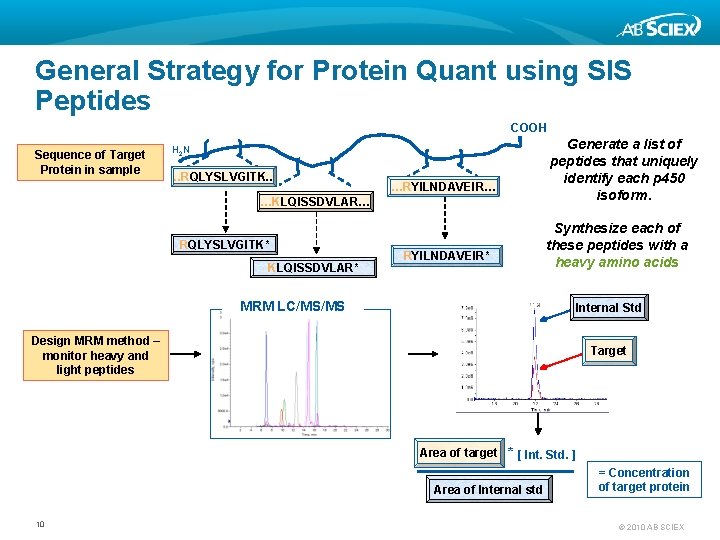
General Strategy for Protein Quant using SIS Peptides COOH Sequence of Target Protein in sample Generate a list of peptides that uniquely identify each p 450 isoform. H 2 N . . RQLYSLVGITK. . …RYILNDAVEIR… …KLQISSDVLAR… RQLYSLVGITK* KLQISSDVLAR* Synthesize each of these peptides with a heavy amino acids RYILNDAVEIR* MRM LC/MS/MS Internal Std Design MRM method – monitor heavy and light peptides Target Area of target * [ Int. Std. ] Area of Internal std 10 = Concentration of target protein © 2010 AB SCIEX

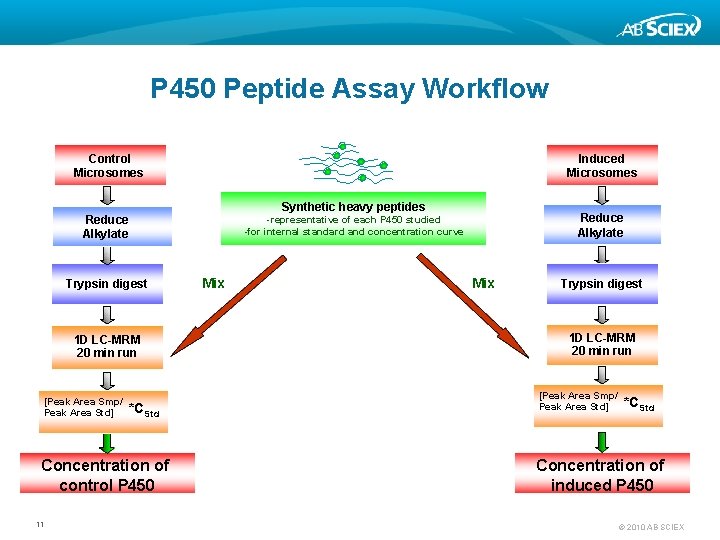
P 450 Peptide Assay Workflow Control Microsomes Induced Microsomes Synthetic heavy peptides Reduce Alkylate Trypsin digest 1 D LC-MRM 20 min run [Peak Area Smp/ Peak Area Std] *CStd Concentration of control P 450 11 Reduce Alkylate -representative of each P 450 studied -for internal standard and concentration curve Mix Trypsin digest 1 D LC-MRM 20 min run [Peak Area Smp/ Peak Area Std] *CStd Concentration of induced P 450 © 2010 AB SCIEX

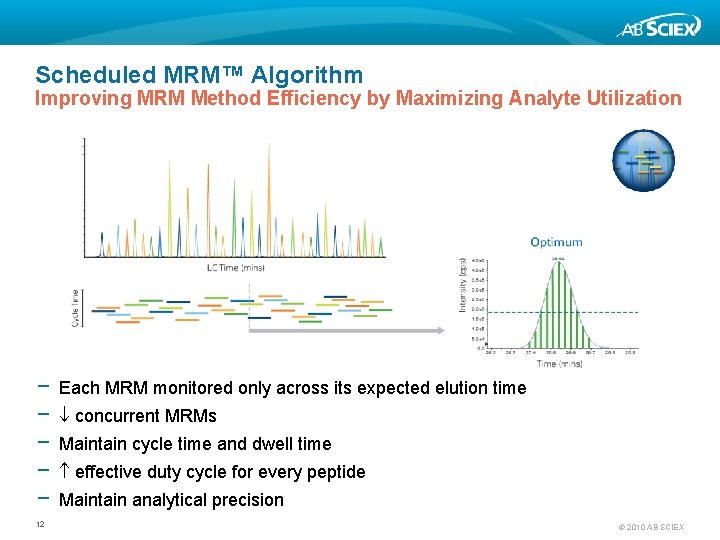
Scheduled MRM™ Algorithm Improving MRM Method Efficiency by Maximizing Analyte Utilization − − − 12 Each MRM monitored only across its expected elution time concurrent MRMs Maintain cycle time and dwell time effective duty cycle for every peptide Maintain analytical precision © 2010 AB SCIEX

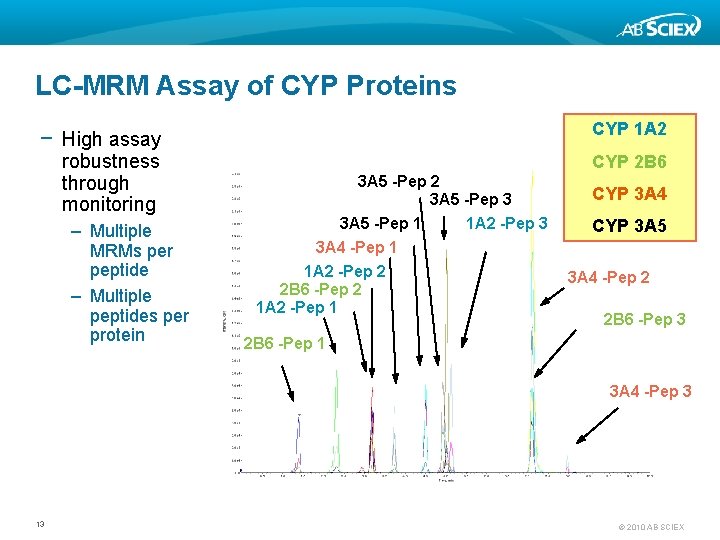
LC-MRM Assay of CYP Proteins CYP 1 A 2 − High assay robustness through monitoring – Multiple MRMs per peptide – Multiple peptides per protein CYP 2 B 6 3 A 5 -Pep 2 3 A 5 -Pep 3 3 A 5 -Pep 1 1 A 2 -Pep 3 3 A 4 -Pep 1 1 A 2 -Pep 2 2 B 6 -Pep 2 1 A 2 -Pep 1 CYP 3 A 4 CYP 3 A 5 3 A 4 -Pep 2 2 B 6 -Pep 3 2 B 6 -Pep 1 3 A 4 -Pep 3 13 © 2010 AB SCIEX

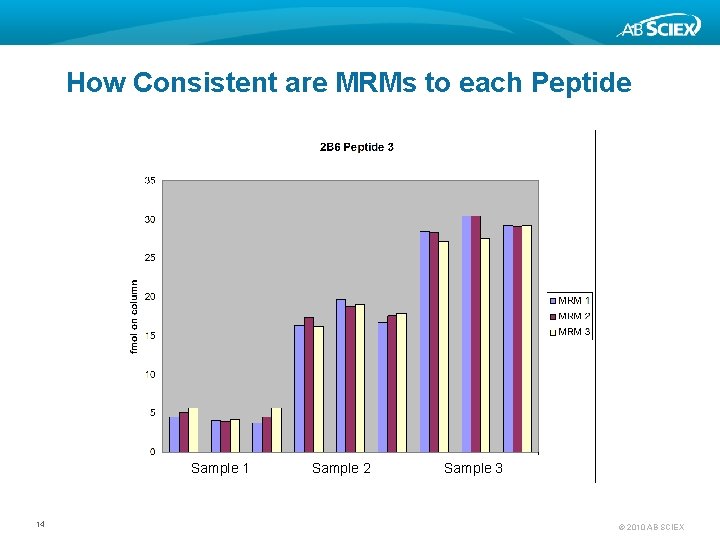
How Consistent are MRMs to each Peptide Sample 1 14 Sample 2 Sample 3 © 2010 AB SCIEX

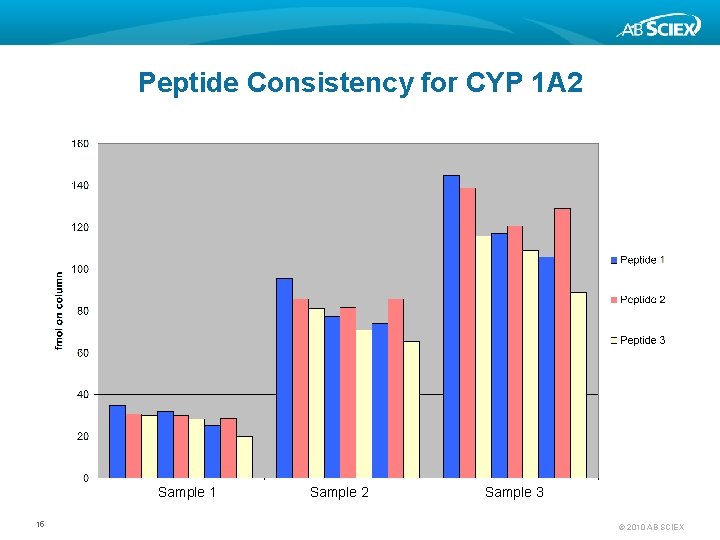
Peptide Consistency for CYP 1 A 2 Sample 1 15 Sample 2 Sample 3 © 2010 AB SCIEX

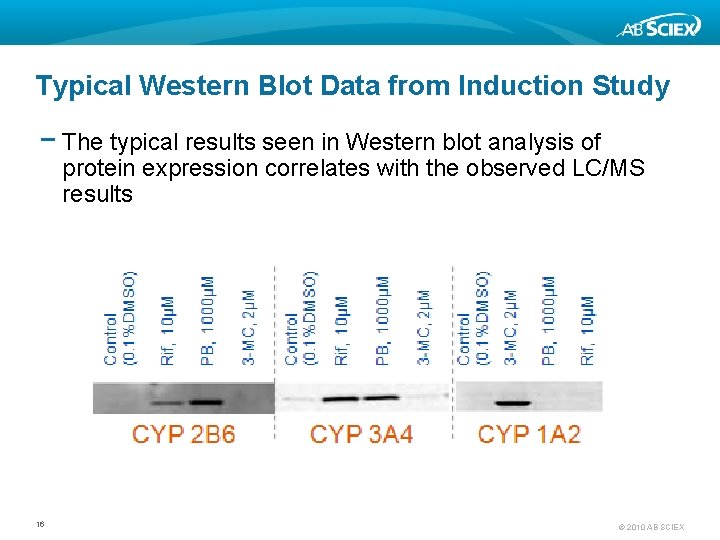
Typical Western Blot Data from Induction Study − The typical results seen in Western blot analysis of protein expression correlates with the observed LC/MS results 16 © 2010 AB SCIEX

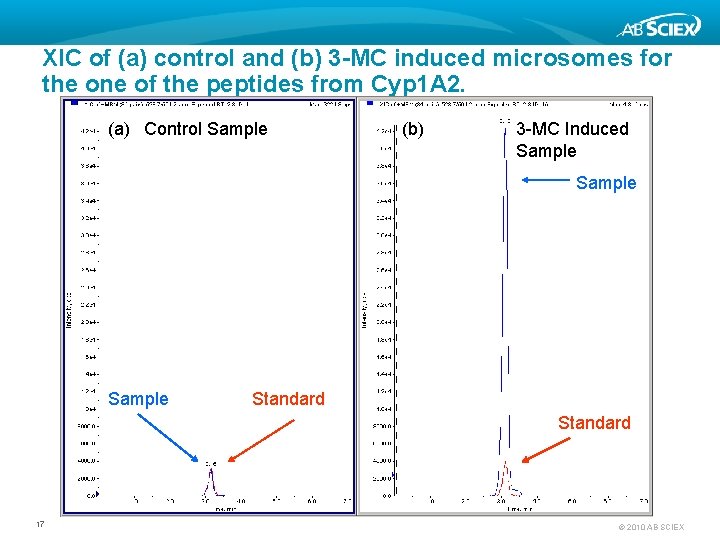
XIC of (a) control and (b) 3 -MC induced microsomes for the one of the peptides from Cyp 1 A 2. (a) Control Sample (b) 3 -MC Induced Sample Standard 17 © 2010 AB SCIEX

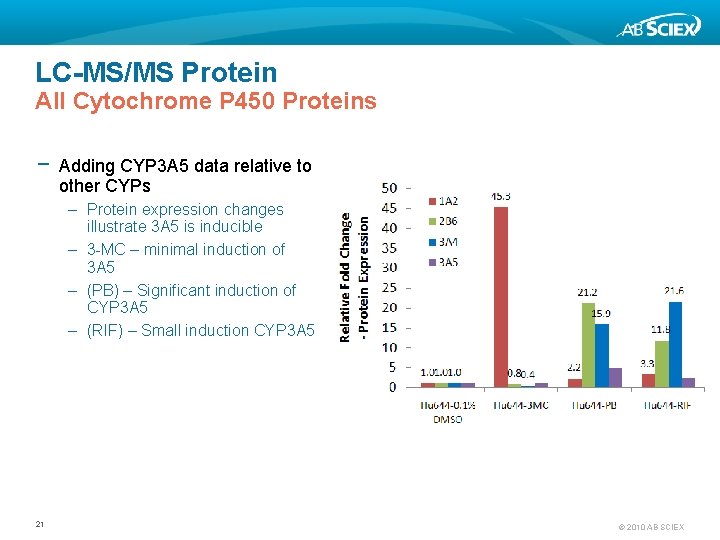
LC-MS/MS Protein All Cytochrome P 450 Proteins − Adding CYP 3 A 5 data relative to other CYPs – Protein expression changes illustrate 3 A 5 is inducible – 3 -MC – minimal induction of 3 A 5 – (PB) – Significant induction of CYP 3 A 5 – (RIF) – Small induction CYP 3 A 5 21 © 2010 AB SCIEX

Conclusions - CYP Induction Assay LC-MS/MS Protein Expression Analysis − Highly sensitive, specific, and fast Multiple Reaction Monitoring (MRM) method has been developed: – 12 different peptides representing 4 unique P 450 proteins (CYP 1 A 2, 2 B 6, 3 A 4 and 3 A 5) were simultaneously monitored and quantified – 2 B 6, a lower abundant CYP, is easily detected showing good dynamic range of method − Largest protein expression change was observed for microsomes prepared from RIF induced hepatocytes – Cyp 3 A 4 showed an increase in expression upon drug treatment of ~50 -fold over control. – S 9 or microsomal subcellular fractions can be used − This method was in excellent agreement with existing methods (m. RNA, enzyme activity assays) 23 © 2010 AB SCIEX

Human Induction Kit (100 Assays) Starter Kit Contents − Heavy peptide mix − Denaturant, Reducing reagent, Alkylating reagent − Digestion buffer − Trypsin − Peptide column − Acquisitions methods for – AB SCIEX Triple Quad™ 5500 and QTRAP® 5500 systems – API 4000™ system, 4000 QTRAP® system, API 5000™ system − Quantitation methods for Multi. Quant™ software 1. 2 − Microsoft Excel 2007 results template 24 © 2010 AB SCIEX

Acknowledgements − AB SCIEX – Sean Seymour – Christie Hunter – Lydia Nuwaysir − Cellz. Direct – Jeanette Hill – Rob Taylor 25 © 2010 AB SCIEX

Thank You for your Attention

Trademarks/Licensing − For Research Use Only. Not for use in diagnostic procedures. − The trademarks mentioned herein are the property of AB Sciex Pte. Ltd. or their respective owners. AB SCIEX™ is being used under license. − © 2010 AB SCIEX. All rights reserved. Information subject to change without notice. 27 © 2010 AB SCIEX