Quantitative Gene Expression Analysis using Microarray and q





- Slides: 35

Quantitative Gene Expression Analysis using Microarray and q. RT-PCR Prof. Jiang B. Liu Computer Science & Information Systems Department Bradley University Peoria, Illinois 61625, U. S. A. BRADLEY U N I V E R S I T Y

TOPICS – Normalization of Micro. Array Data – Genomic Data Analysis using q. RT-PCR – Pathway Analysis – Conclusion Quantitative Genomic Data Analysis BRADLEY U N I V E R S I T Y

1. Normalization of Microarray Data • We have worked with USDA National Center for Agriculture Utilization Research (ARS) on developing external RNA controls for quantitative gene expression analysis on both microarray and Real time quantitative reverse transcription polymerase chain reaction (q. RT-PCR) platforms. • Normalization of microarray data is a vital important step in analyze the generated raw data by eliminating or reduce the effects which arise from variation in the technology rather than from biological differences between the RNA samples or between the printed probes. Quantitative Genomic Data Analysis BRADLEY U N I V E R S I T Y

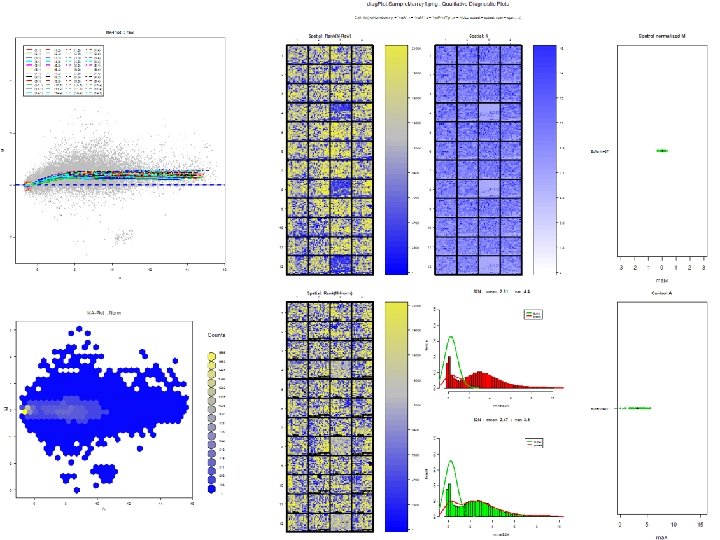
Normalization of Microarray Data • For microarray data acquisition, microarray slides were processed using Gene. Pix 4000 B scanner. Data files were generated using Gene. Pix Pro software. • Several software tools were investigated for providing an environment for analyzing both transcriptone data (gene expression data) and metabolome data (ompound data). Microarray data analysis has been attempted using Bioconductor R software and KEGG Keg. Array software. Quantitative Genomic Data Analysis BRADLEY U N I V E R S I T Y

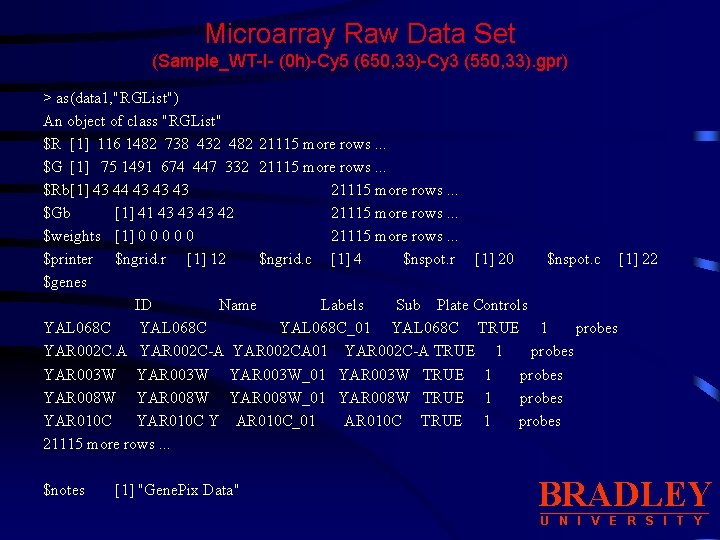
Microarray Raw Data Set (Sample_WT-I- (0 h)-Cy 5 (650, 33)-Cy 3 (550, 33). gpr) > as(data 1, "RGList") An object of class "RGList" $R [1] 116 1482 738 432 482 21115 more rows. . . $G [1] 75 1491 674 447 332 21115 more rows. . . $Rb[1] 43 44 43 43 43 21115 more rows. . . $Gb [1] 41 43 43 43 42 21115 more rows. . . $weights [1] 0 0 0 21115 more rows. . . $printer $ngrid. r [1] 12 $ngrid. c [1] 4 $nspot. r [1] 20 $nspot. c [1] 22 $genes ID Name Labels Sub Plate Controls YAL 068 C_01 YAL 068 C TRUE 1 probes YAR 002 C. A YAR 002 C-A YAR 002 CA 01 YAR 002 C-A TRUE 1 probes YAR 003 W_01 YAR 003 W TRUE 1 probes YAR 008 W_01 YAR 008 W TRUE 1 probes YAR 010 C Y AR 010 C_01 AR 010 C TRUE 1 probes 21115 more rows. . . $notes [1] "Gene. Pix Data" BRADLEY U N I V E R S I T Y

BRADLEY U N I V E R S I T Y

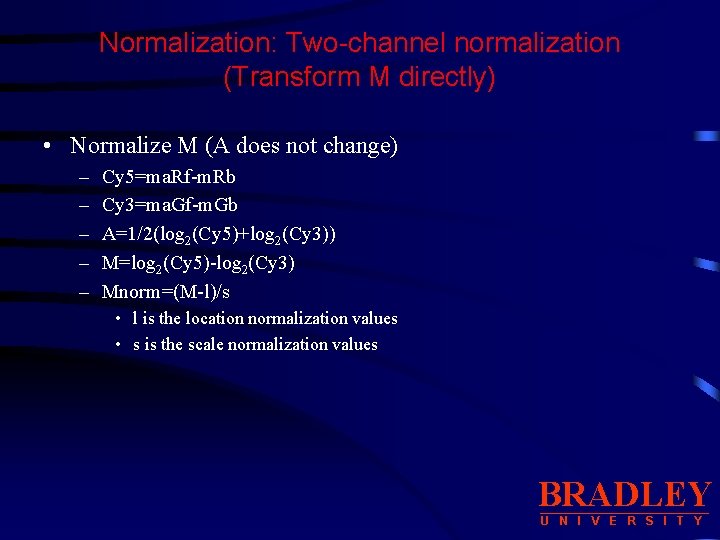
Normalization: Two-channel normalization (Transform M directly) • Normalize M (A does not change) – – – Cy 5=ma. Rf-m. Rb Cy 3=ma. Gf-m. Gb A=1/2(log 2(Cy 5)+log 2(Cy 3)) M=log 2(Cy 5)-log 2(Cy 3) Mnorm=(M-l)/s • l is the location normalization values • s is the scale normalization values BRADLEY U N I V E R S I T Y

a. Location Normalization Procedure: print. Tip. Loess (default) • print. Tip. Loess: for within-print-tip-group intensity dependent location normalization using the loess (Local Polynomial Regression Fitting) function. normdata<-ma. Norm. Main(mbatch = mbatch, f. loc = list(ma. Norm. Loess(x = "ma. A", y = "ma. M", z = "ma. Print. Tip", w = NULL, subset = subset, span = span, . . . )), Mloc = Mloc, Mscale = Mscale, echo = echo) BRADLEY U N I V E R S I T Y

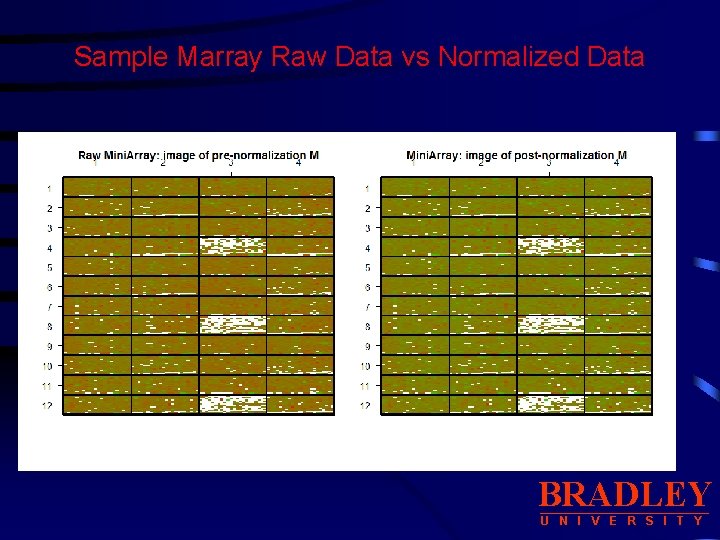
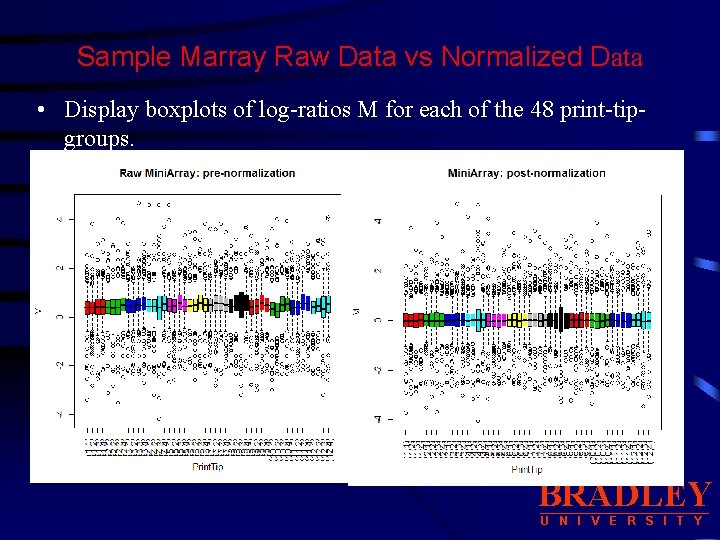
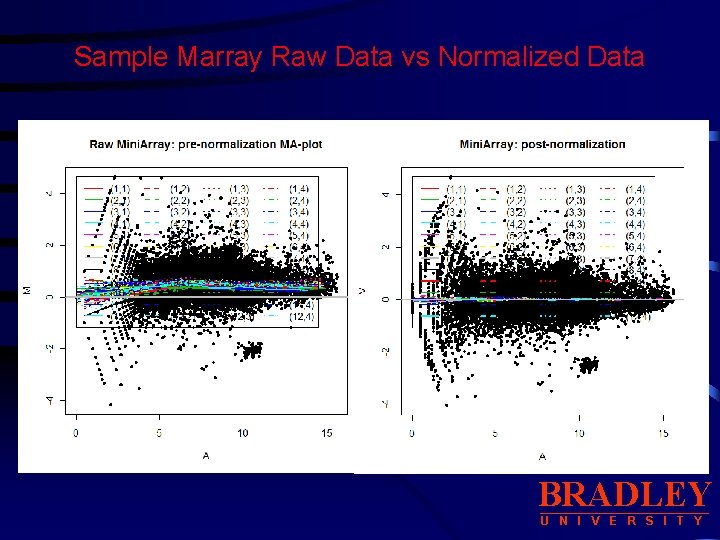
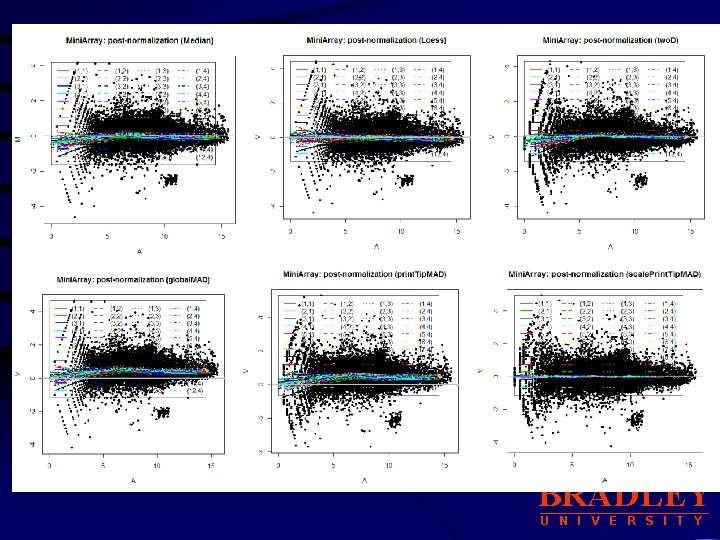
Sample Marray Raw Data vs Normalized Data BRADLEY U N I V E R S I T Y

Sample Marray Raw Data vs Normalized Data • Display boxplots of log-ratios M for each of the 48 print-tipgroups. BRADLEY U N I V E R S I T Y

Sample Marray Raw Data vs Normalized Data BRADLEY U N I V E R S I T Y

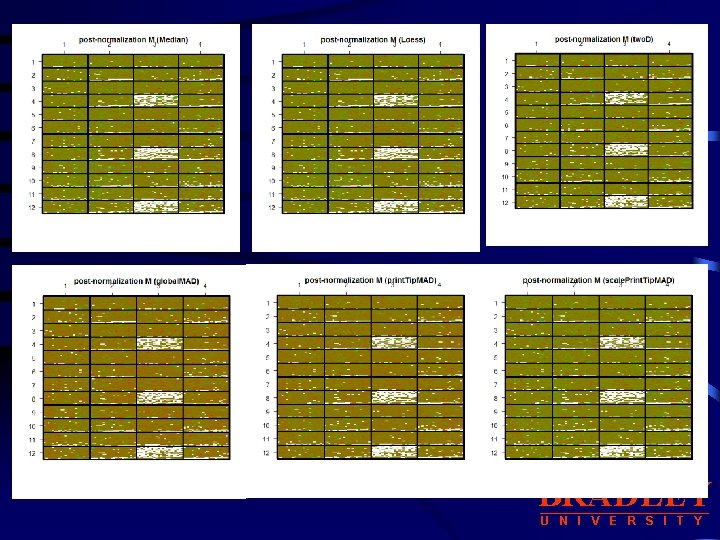
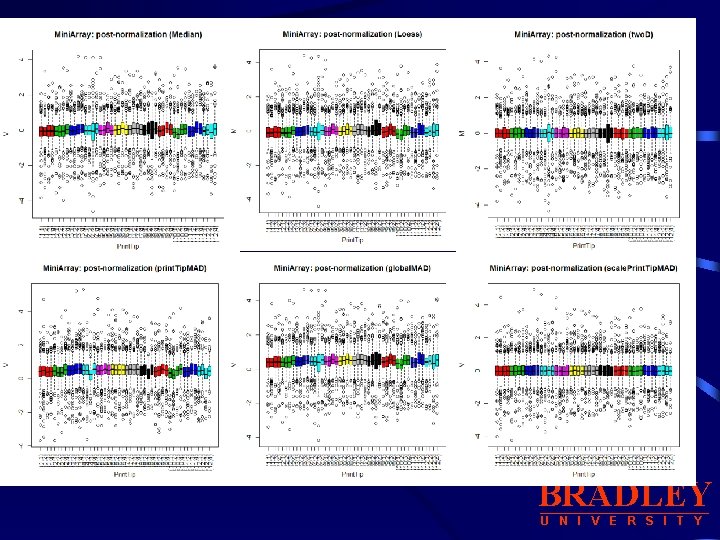
b. Other Location, Scale Normalization Procedures: median, loess, two. D, Print. Tip. MAD, global. MAD, scale. Print. Tip. MAD • median: for global median location normalization • loess: for global intensity or A-dependent location normalization using the loess function. • two. D: for 2 D spatial location normalization using the loess function • global. MAD: for global scale normalization using the median absolute deviation (MAD), this allows between slide scale normalization. • print. Tip. MAD: for within-print-tip-group scale normalization using the • median absolute deviation (MAD). This argument can be specified the first letter of each method. using • scale. Print. Tip. MAD : for within-print-tip-group intensity dependent location normalization followed by within-print-tip-group scale normalization using the median absolute deviation (MAD). BRADLEY U N I V E R S I T Y

BRADLEY U N I V E R S I T Y

BRADLEY U N I V E R S I T Y

BRADLEY U N I V E R S I T Y

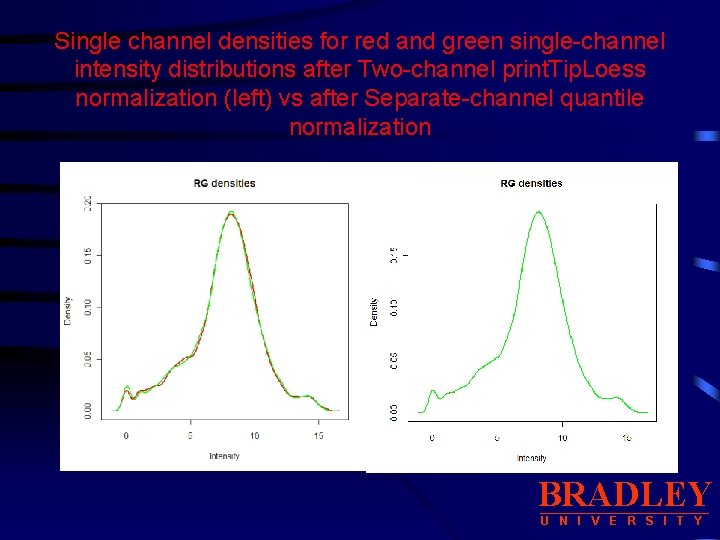
Normalization: Separate-channel normalization limma package: quantile • Normalize the red and green absolute intensities separately. • Use quantile method (on ma. Norm data): • normdata. p<-as(normdata, ”MAList”) • normdata. pq<-normalize. Between. Arrays(normdata. p, method=“quantile”) • plot. Densities(normdata. pq) BRADLEY U N I V E R S I T Y

Single channel densities for red and green single-channel intensity distributions after Two-channel print. Tip. Loess normalization (left) vs after Separate-channel quantile normalization BRADLEY U N I V E R S I T Y

2. Genomic Data Analysis Using q. RT-PCR • The conventional normalizations use the housekeeping genes as an internal reference. With increased concerns of variability of housekeeping genes in response to varies conditions, there is no commonly accepted housekeeping genes (Salmon vs. Bean). • The q. RT-PCR has been accepted recently as the assay choice for m. RNA quantification. We have worked on several q. RTPCR data analysis methods for a deeper understanding of the raw data with the control genes as an external reference and target genes expression and the relationships among them. All complex interactions of the organism information were analyzed using both the statistical and pathway gene expression methods. Quantitative Genomic Data Analysis BRADLEY U N I V E R S I T Y

Genomic Data Analysis Using q. RT-PCR • To analyze the q. RT-PCR data, it is important to catch and eliminate the non-biological variation across the range. • Because the control genes are affected by all sources of variation during the experimental workflow in the same way as the target genes, the current golden standard for q. RT-PCR data normalization is to use the control genes to normalize the target genes. The selected control genes must be validated to make sure that they are stable. Once a set of control genes are selected and validated, then they can be used to generate the normalize factor for the target genes normalization, since they are measured using the same methodology. (We used control genes as the external references) Quantitative Genomic Data Analysis BRADLEY U N I V E R S I T Y

Genomic Data Analysis Using q. RT-PCR • Currently there is no universally accepted method for data normalization for q. RT-PCR data. We can compare several existed methods used with the USDA-ARS data and develop new methods that fit well with our q. RT-PCR experiment. • After data validation, we have developed an automatic process for raw data handling using a house developed software. The executable Masterq. RT-PCR. exe program is used to compute the master linear regression equation and associate statistics based on the control genes as an external reference and to generate the estimate m. RNA value of all target genes and copy number for each target gene entered by the user. Quantitative Genomic Data Analysis BRADLEY U N I V E R S I T Y

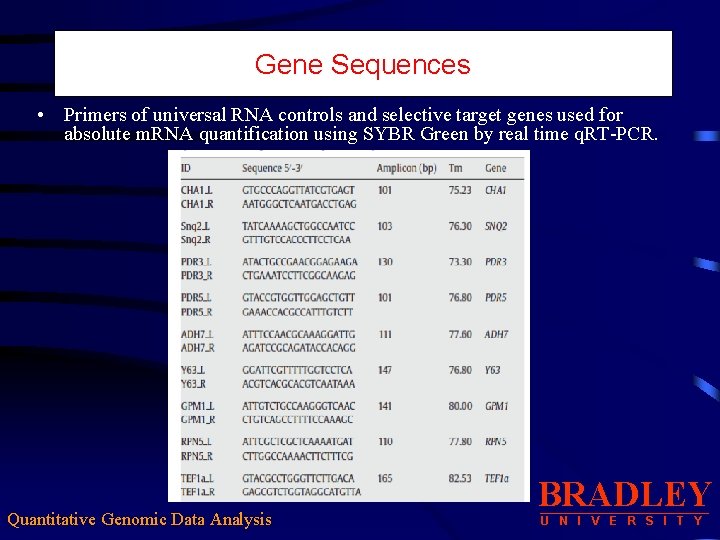
Gene Sequences • Primers of universal RNA controls and selective target genes used for absolute m. RNA quantification using SYBR Green by real time q. RT-PCR. Quantitative Genomic Data Analysis BRADLEY U N I V E R S I T Y

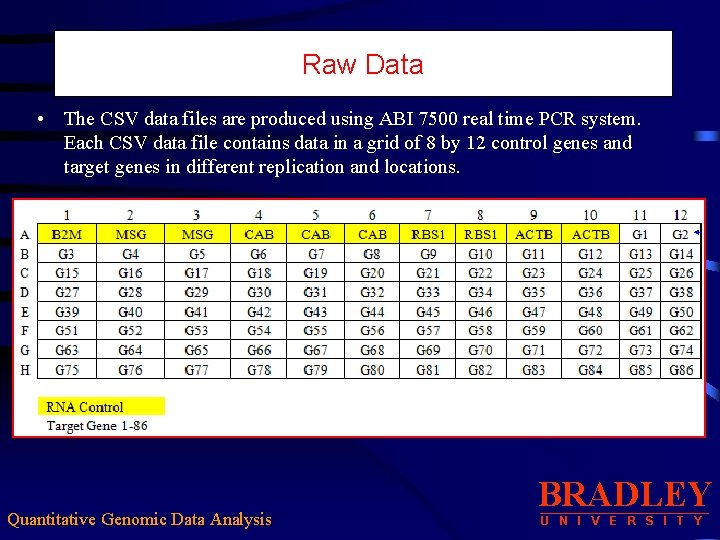
Raw Data • The CSV data files are produced using ABI 7500 real time PCR system. Each CSV data file contains data in a grid of 8 by 12 control genes and target genes in different replication and locations. Quantitative Genomic Data Analysis BRADLEY U N I V E R S I T Y

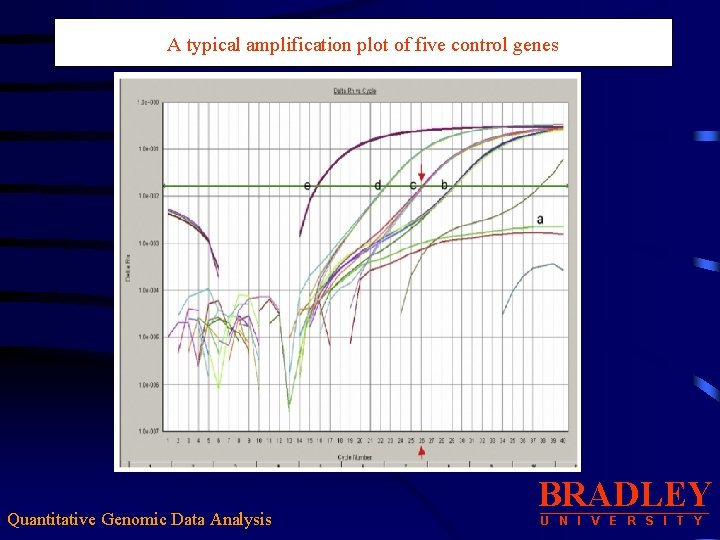
A typical amplification plot of five control genes Quantitative Genomic Data Analysis BRADLEY U N I V E R S I T Y

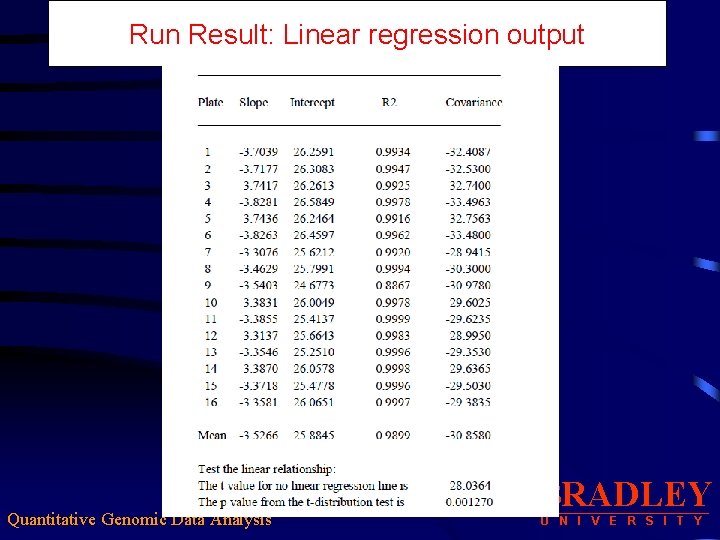
Run Result: Linear regression output Quantitative Genomic Data Analysis BRADLEY U N I V E R S I T Y

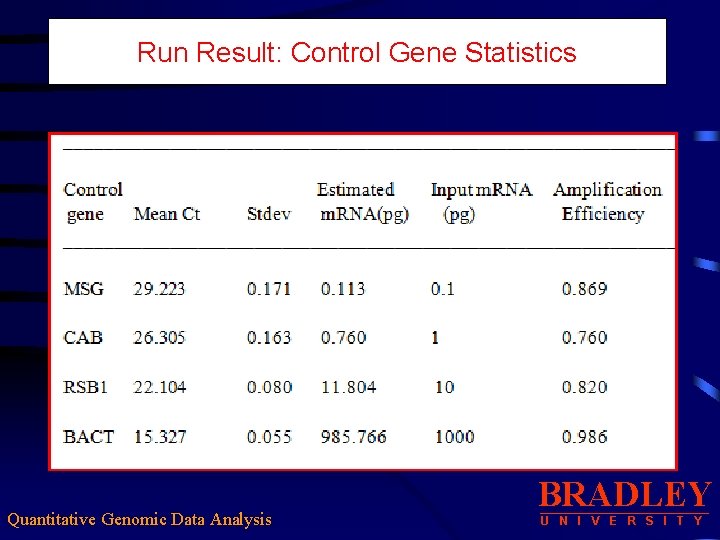
Run Result: Control Gene Statistics Quantitative Genomic Data Analysis BRADLEY U N I V E R S I T Y

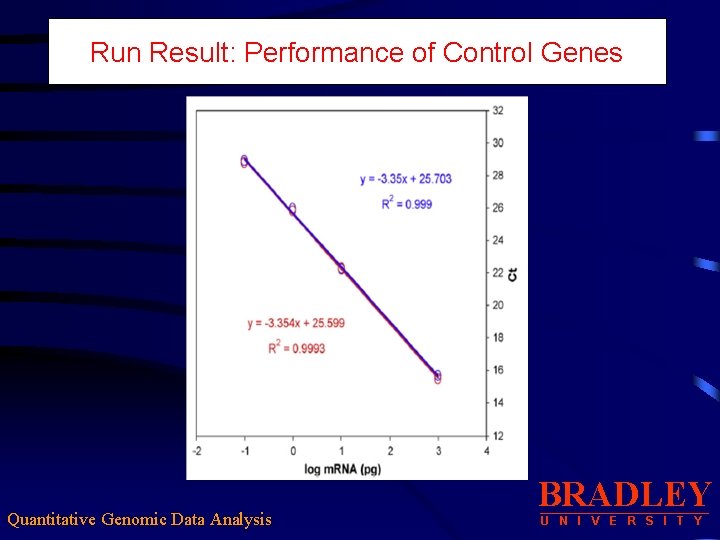
Run Result: Performance of Control Genes Quantitative Genomic Data Analysis BRADLEY U N I V E R S I T Y

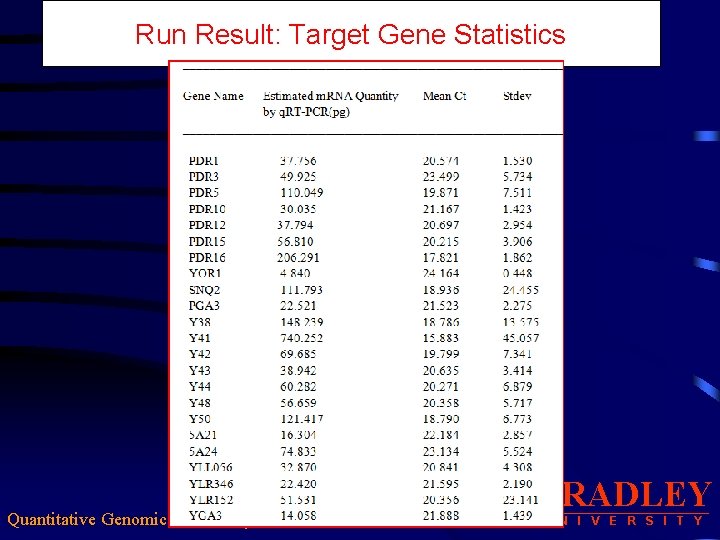
Run Result: Target Gene Statistics Quantitative Genomic Data Analysis BRADLEY U N I V E R S I T Y

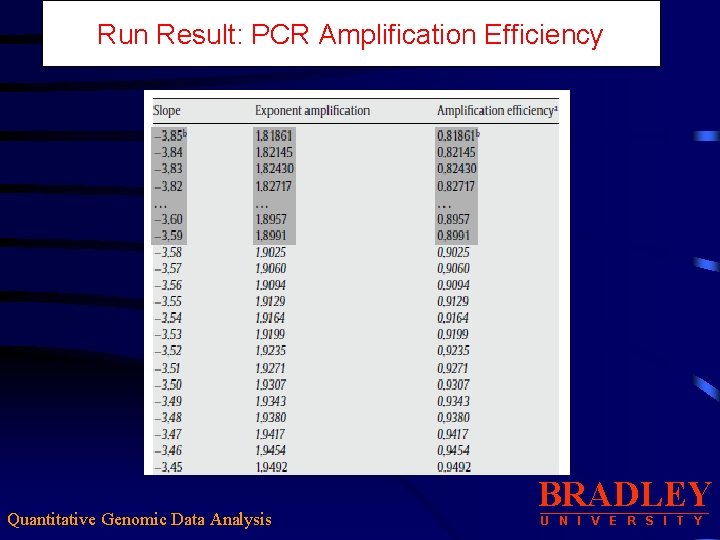
Run Result: PCR Amplification Efficiency Quantitative Genomic Data Analysis BRADLEY U N I V E R S I T Y

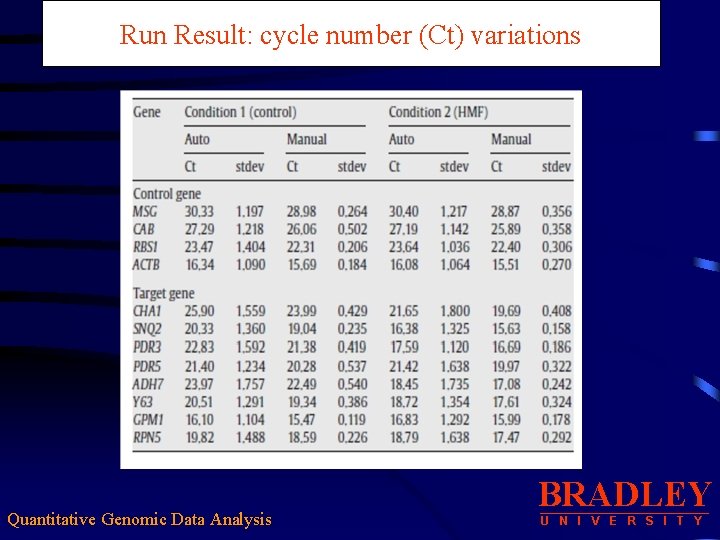
Run Result: cycle number (Ct) variations Quantitative Genomic Data Analysis BRADLEY U N I V E R S I T Y

Normalization using the Developed Control System • The unique control system developed for real time PCR data has four reference genes that were selected to generated master regression relationship for other genes of interest, i. e. target genes. We can further look at the normalization of the target genes using the selected control genes and develop a more comprehensive program using the robust control system. 1) Validate the Control genes for the control systems developed. The four control genes, MSG, CAB, RBS 1, and ACTB, are used to generate the control system for the other target genes. The range of these control genes covers the whole range of the target genes. The stability of these genes can further be validated either by statistical or biological methods. Quantitative Genomic Data Analysis BRADLEY U N I V E R S I T Y

Normalization using the Developed Control System • Method 1: Select and validate control genes using M-value Select control genes belong to different functional and abundance classes to reduce the risk that genes are co-regulated. Select at least three control genes, with 4 th or 5 th control gene to deal with the observed expression variation. • Method 2: Select and validate control genes using variance of Ct value Use Ct value instead of relative quantities. Remove outlier value gene based on the variation of Ct value. • Method 3: Select and validate control genes using advanced statistical models Rank the control genes using statistical formula. Estimate the variance of the log-average of the repression of control genes. Conduct t-test or F-test to evaluate the control genes • Method 4: Select and validate control genes using Biological testing Quantitative Genomic Data Analysis BRADLEY U N I V E R S I T Y

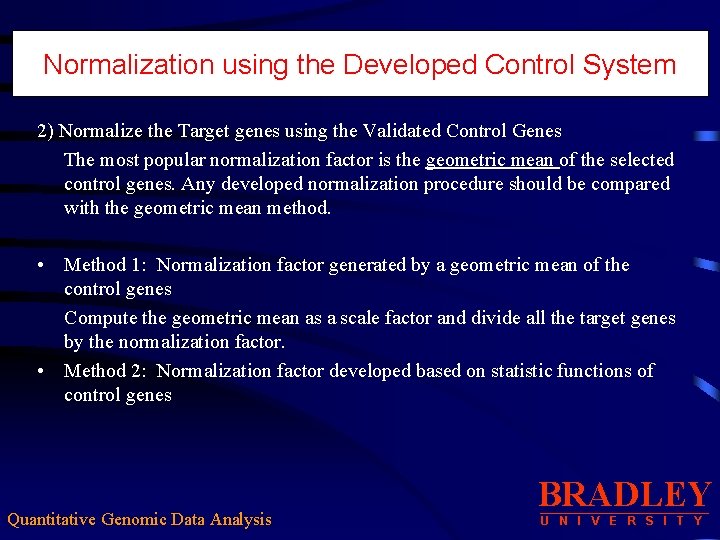
Normalization using the Developed Control System 2) Normalize the Target genes using the Validated Control Genes The most popular normalization factor is the geometric mean of the selected control genes. Any developed normalization procedure should be compared with the geometric mean method. • Method 1: Normalization factor generated by a geometric mean of the control genes Compute the geometric mean as a scale factor and divide all the target genes by the normalization factor. • Method 2: Normalization factor developed based on statistic functions of control genes Quantitative Genomic Data Analysis BRADLEY U N I V E R S I T Y

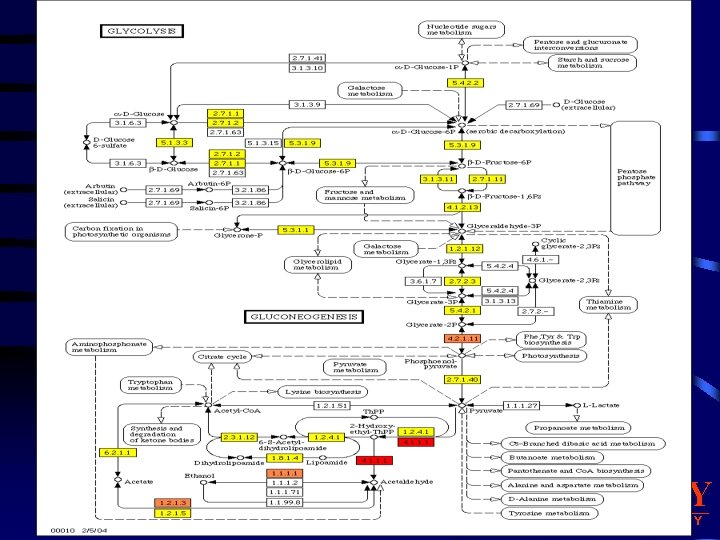
3. Pathway Analysis Normalized and validated genomic data from our control system can then be used to generate Pathway maps. • PATHWAY maps representing our knowledge on the molecular interaction and reaction networks for: 1. Metabolism 2. Genetic Information Processing 3. Environmental Information Processing 4. Cellular Processes 5. Human Diseases 6. Drug Development. We have design a My. SQL database to store the USDA-ARS experimental genomic data, a Perl software package to load the data into the tables and generate the pathway diagrams from KEGG pathway database. Quantitative Genomic Data Analysis BRADLEY U N I V E R S I T Y

BRADLEY U N I V E R S I T Y

Conclusions • We have worked with USDA-ARS on Microarray and q. RT-PCR genomic data analysis and developed a control system and associated software package to process the data. – Journal of Biotechnology. • The robust standard reference for absolute m. RNA quantification using q. RT-PCR control system provides a reliable reference for absolute quantification of m. RNA using the q. RT-PCR assay and simplifies the conventional q. RT-PCR procedures, and increased data reliability, reproducibility, and throughput of the assay. • The target genes generated from the control genes can then be used for further genomic data analysis such as Pathway analysis for applications such as Genetic Information Processing. Quantitative Genomic Data Analysis BRADLEY U N I V E R S I T Y