Quality of services of a primary nucleotide sequence















- Slides: 30

Quality of services of a primary nucleotide sequence database Hideaki Sugawara Center for Information Biology and DDBJ, National Institute of Genetics SOKEN-DAI. 2006/10/23 Primary Biological Databases

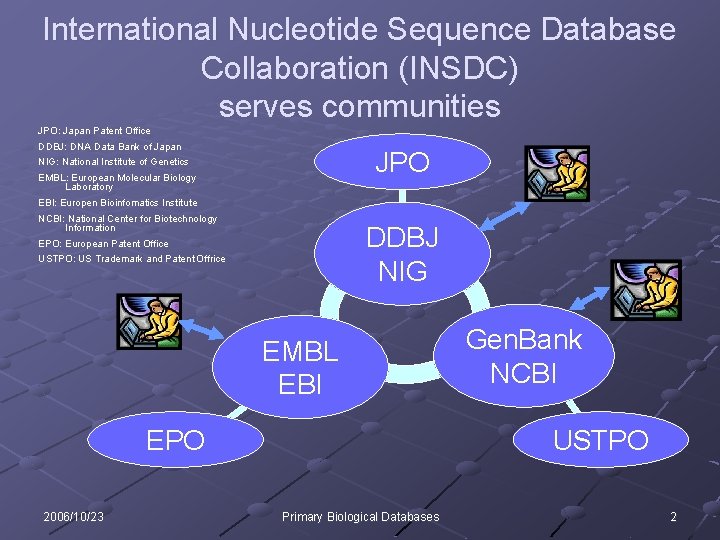
International Nucleotide Sequence Database Collaboration (INSDC) serves communities JPO: Japan Patent Office DDBJ: DNA Data Bank of Japan JPO NIG: National Institute of Genetics EMBL: European Molecular Biology Laboratory EBI: Europen Bioinfomatics Institute NCBI: National Center for Biotechnology Information DDBJ NIG EPO: European Patent Office USTPO: US Trademark and Patent Offrice EMBL EBI EPO 2006/10/23 Gen. Bank NCBI USTPO Primary Biological Databases 2

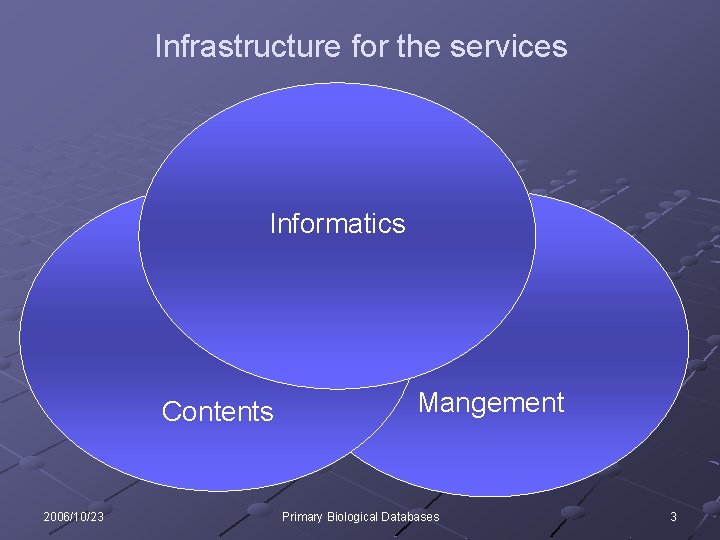
Infrastructure for the services Informatics Contents 2006/10/23 Mangement Primary Biological Databases 3

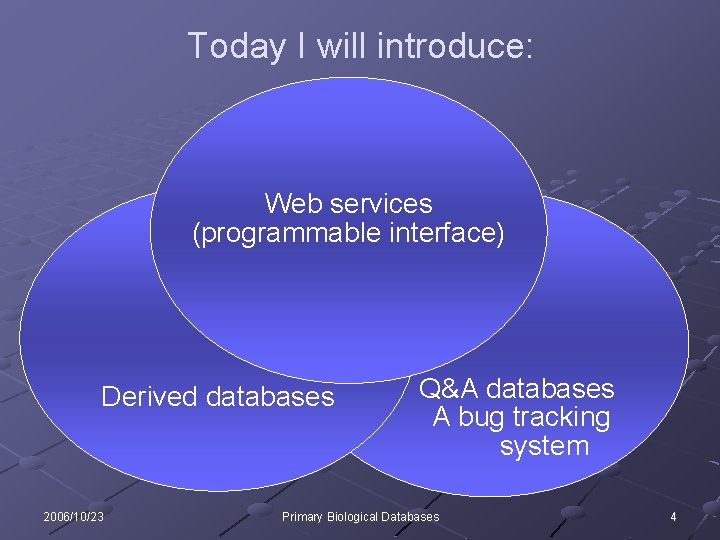
Today I will introduce: Web services (programmable interface) Derived databases 2006/10/23 Q&A databases A bug tracking system Primary Biological Databases 4

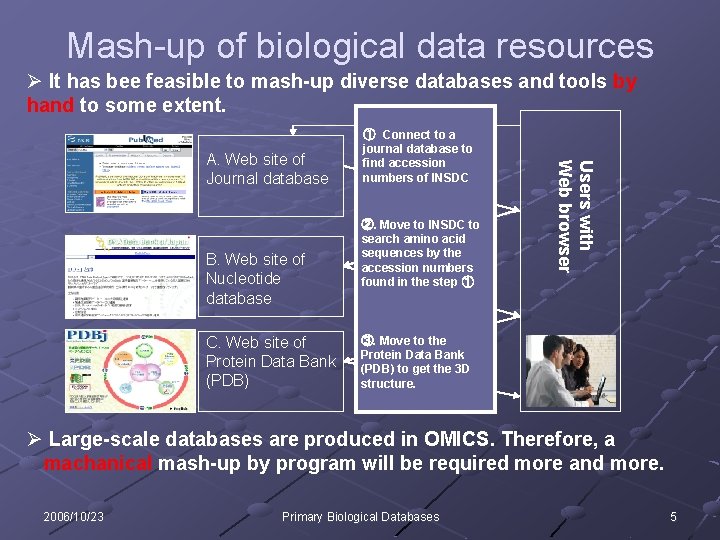
Mash-up of biological data resources Ø It has bee feasible to mash-up diverse databases and tools by hand to some extent. B. Web site of Nucleotide database C. Web site of Protein Data Bank (PDB) ②. Move to INSDC to search amino acid sequences by the accession numbers found in the step ① Users with Web browser A. Web site of Journal database ① Connect to a journal database to find accession numbers of INSDC ③. Move to the Protein Data Bank (PDB) to get the 3 D structure. Ø Large-scale databases are produced in OMICS. Therefore, a machanical mash-up by program will be required more and more. 2006/10/23 Primary Biological Databases 5

Web services for the biological problem solving environment We implemented a SOAP (Simple Object Access Protocol) server and Web services that provide a program-friendly interface. We propose standardization of bioinformatics services to improve the interoperability of desperately diverse biological data resources. 2006/10/23 Primary Biological Databases 6

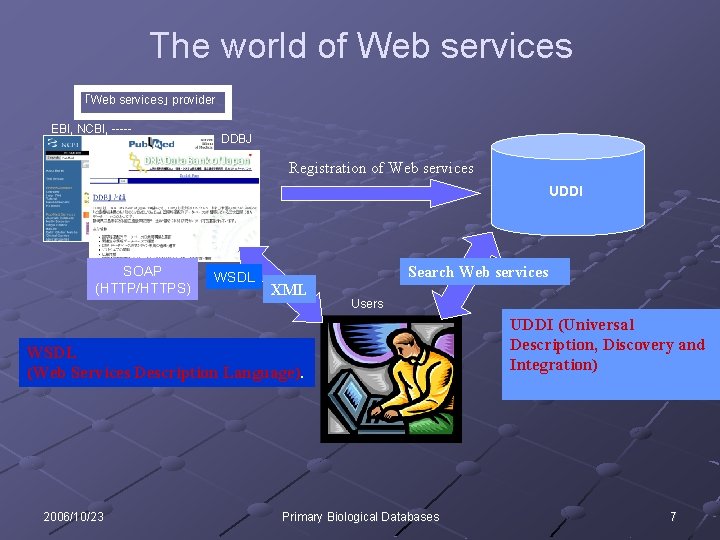
The world of Web services 「Web services」 provider EBI, NCBI, ----- DDBJ Registration of Web services UDDI SOAP (HTTP/HTTPS) WSDL XML Search Web services Users WSDL (Web Services Description Language). 2006/10/23 Primary Biological Databases UDDI (Universal Description, Discovery and Integration) 7

XML Central of DDBJ: http: //xml. nig. ac. jp/index. html 2006/10/23 Primary Biological Databases 8

Workflow composed of methods provided by Web services 141 methods in 15 services are available. Blast Clustal. W DDBJ Ensembl Fasta Getentry GIB GTOP NCBI Genome Annotation PML Ref. Seq SPS SRS Tx. Search Vec. Screen OMIM Gene Ontology Rfam Open developed Custom made workflows 2006/10/23 Primary Biological Databases 9

Easy to bind Web services methods from your program Ø Download Active. Perl for WINDOWS and you can call methods in DDBJ Web services. Ø Specify a WDSL file and call a method: In the following example, the method of get. XML_DDBJEntry(accession) of the Getentry Web services is called from a Perl program: 1. Include the package needed to use SOAP service. 2. Specifies WSDL file of SOAP service you want to use. 3. Call the service you want to use. 2006/10/23 Primary Biological Databases 10

Workflow with Online Mendelian Inheritance in Man (OMIM) Web services tool Data Local program This workflow reveal homology relationship between human disease genes and genes of other eukaryotes. 2006/10/23 Primary Biological Databases 11

Environmental DNA sequences automatic annotation workflow In order to make full use of gene information included in nucleotide sequence database, we developed workflow of gene finding of DNA fragments obtained from pooled genome samples of uncultured microbes in environmental samples. BLASTP Environmental Sample EMBOSS (getorf) ORF Inter. Pro Scan Selected ORF Attached Product FLOW 1. Execute getorf in EMBOSS which finds and outputs the sequences of open reading frames (ORFs). The ORFs can be defined as regions of a specified minimum size (longer than 300 bp) between START and STOP codons. (The minimum size cut-off for getorf was ) 2. The protein product was attacheded by searching the bacterial division of DDBJ release 62 against predicted ORFs using BLASTP. (The e-value cut-off for BLASTP was 1 e-40. ) 2006/10/23 Primary Biological Databases 12

Access frequency of DDBJ Web services • Examples of methods frequently used: getentry, blast, Ref. Seq、GO 、 2006/10/23 Primary Biological Databases 13

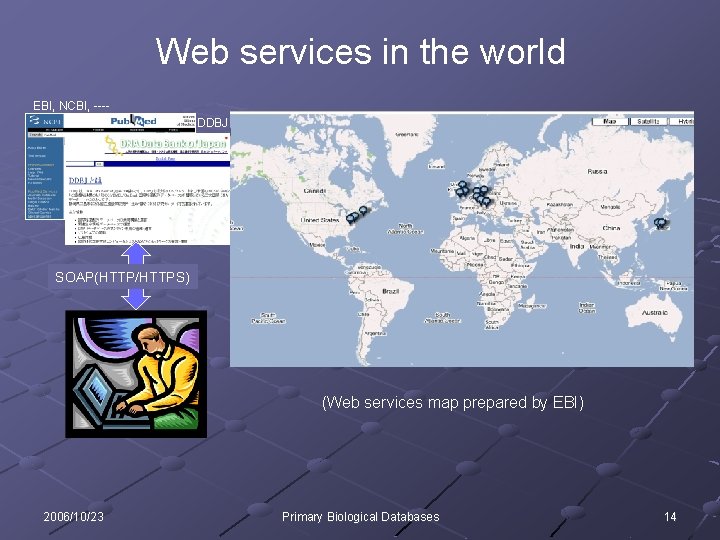
Web services in the world EBI, NCBI, ---DDBJ SOAP(HTTP/HTTPS) 利用者 (Web services map prepared by EBI) 2006/10/23 Primary Biological Databases 14

Derived databases 2006/10/23 Primary Biological Databases 15

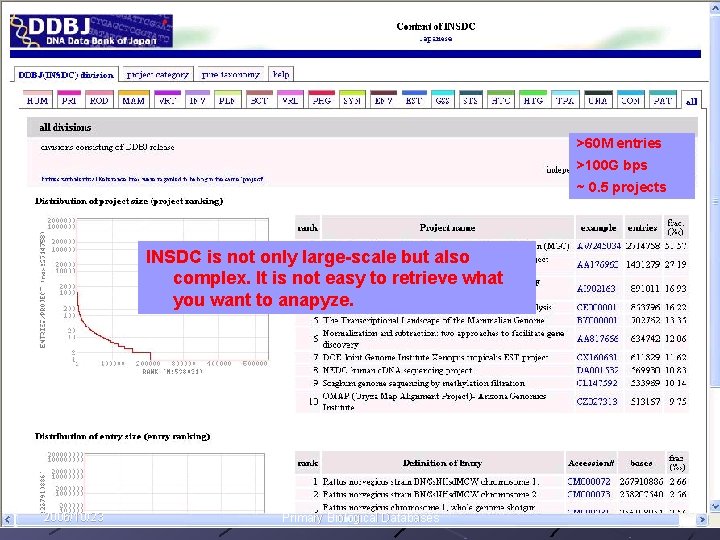
>60 M entries >100 G bps ~ 0. 5 projects INSDC is not only large-scale but also complex. It is not easy to retrieve what you want to anapyze. 2006/10/23 Primary Biological Databases

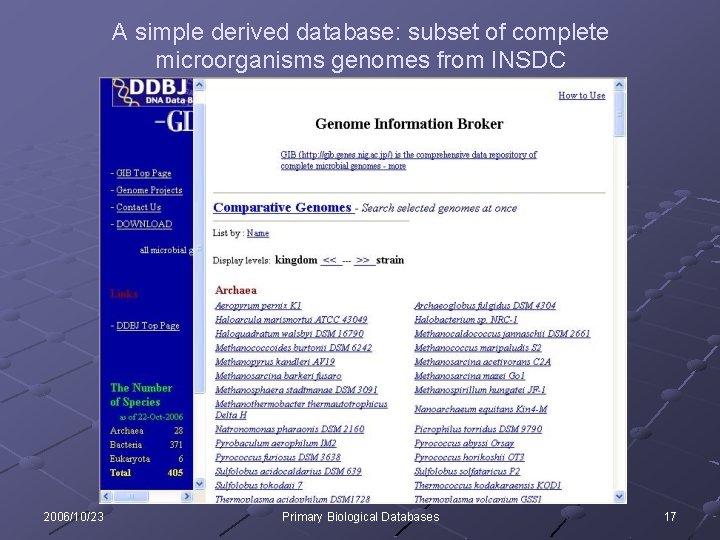
A simple derived database: subset of complete microorganisms genomes from INSDC 2006/10/23 Primary Biological Databases 17

Gene prediction programs used 2006/10/23 Primary Biological Databases 18

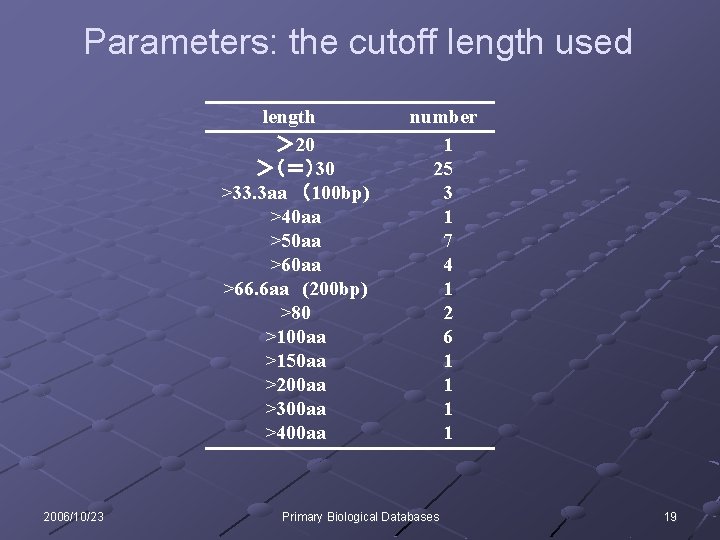
Parameters: the cutoff length used length > 20 >(=)30 >33. 3 aa (100 bp) >40 aa >50 aa >66. 6 aa (200 bp) >80 >100 aa >150 aa >200 aa >300 aa >400 aa 2006/10/23 number 1 25 3 1 7 4 1 2 6 1 1 Primary Biological Databases 19

Annotation: description of products ~ Hahella chejuensis KCTC 2396 CDS 1023521. . 1024429 /gene="arg. B" /locus_tag="HCH_01027" COG ID in the /EC_number="2. 7. 2. 8" ~ Archaeoglobus fulgidus DSM 4304 qualifier of /note="COG 0548" CDS complement(1141715. . 1142587) /codon_start=1 /locus_tag="AF_1280" /transl_table=11 /note="similar to GB: L 77117 SP: Q 60382 PID: 1592260 percent /product="Acetylglutamate kinase"by sequence similarity; identity: 56. 06; identified putative" Some details of/protein_id="ABC 27909. 1" ~ Agrobacterium tumefaciens C 58 circular chromosome /db_xref="GI: 83631942" /codon_start=1 homology CDS complement(373582. . 374466) /translation="MLDRDNALQVAAVLSKALPYIQRFAGKTIVIKYGGNAMTDEELK /transl_table=11 analysis in the. NSFARDVVMMKLVGINPIVVHGGGPQIGDLLQRLNIKSSFINGLRVTDSETMDVVEMV /gene="AGR_C_666" /product="acetylglutamate kinase (arg. B)" /note="acetylglutamate kinase PA 5323 {imported} LGGSVNKDIVALINRNGGKAIGLTGKDANFITARKLEVTRATPDMQKPEIIDIGHVGE /note qualifier /protein_id="AAB 89966. 1" Pseudomonas aeruginosa (strain PAO 1)" VTGVRKDIITMLTDSDCIPVIAPIGVGQDGASYNINADLVAGKVAEVLQAEKLMLLTN /db_xref="GI: 2649301" Product name in /codon_start=1 IAGLMNKEGKVLTGLSTKQVDELIADGTIHGGMLPKIECALSAVKNGVHSAHIIDGRV /translation="MENVELLIEALPYIKDFHSTTMVIKIGGHAMVNDRILEDTIKDI /transl_table=11 PHATLLEIFTDEGVGTLITRKGCDDA" VLLYFVGIKPVVVHGGGPEISEKMEKFGLKPKFVEGLRVTDKETMEVVEMVLDGKVNS the qualifier of /product="AGR_C_666 p" KIVTTFIRNGGKAVGLSGKDGLLIVARKKEMRMKKGEEEVIIDLGFVGETEFVNPEII /note /protein_id="AAK 86197. 1" RILLDNGFIPVVSPVATDLAGNTYNLNADVVAGDIAAALKAKKLIMLTDVPGILENPD /db_xref="GI: 15155294" DKSTLISRIRLSELENMRSKGVIRGGMIPKVDAVIKALKSGVERAHIIDGSRPHSILI /translation="MTSSESEIQARLLAQALPFMQKYENKTIVVKYGGHAMGDSTLGK ELFTKEGIGTMVEP" Product ID in the AFAEDIALLKQSGINPIVVHGGGPQIGAMLSKMGIESKFEGGLRVTDAKTVEIVEMVL qualifier of AGSINKEIVALINQTGEWAIGLCGKDGNMVFAEKAKKTVIDPDSNIERVLDLGFVGEV /product VEVDRTLLDLLAKSEMIPVIAPVAPGRDGATYNINADTFAGAIAGALHATRLLFLTDV PGVLDKNKELIKELTVSEARALIKDGTISGGMIPKVETCIDAIKAGVQGVVILNGKTP HSVLLEIFTEGAGTLIVP" 2006/10/23 Primary Biological Databases 20

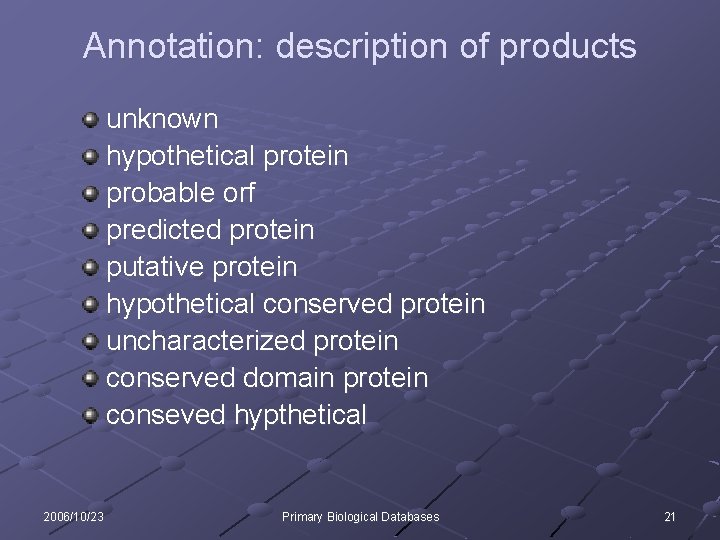
Annotation: description of products unknown hypothetical protein probable orf predicted protein putative protein hypothetical conserved protein uncharacterized protein conserved domain protein conseved hypthetical 2006/10/23 Primary Biological Databases 21

Annotation: inconsistency or biological variation Agrobacterium tumefaciens C 58 circular chromosome (Cereon) Agrobacterium tumefaciens C 58 circular chromosome (U. Washington) 2006/10/23 Primary Biological Databases 22

A derived database: contents of complete microorganisms genomes were evaluated Gene Trek in Procayote Space 2006/10/23 Primary Biological Databases 23

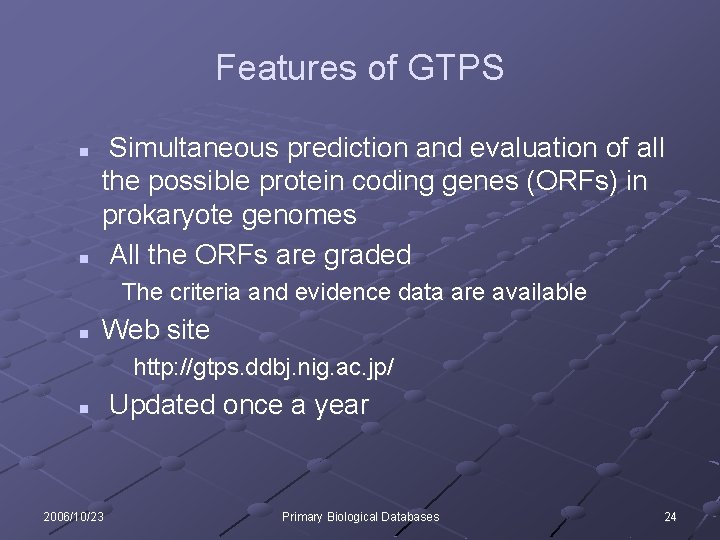
Features of GTPS n n Simultaneous prediction and evaluation of all the possible protein coding genes (ORFs) in prokaryote genomes All the ORFs are graded The criteria and evidence data are available n Web site http: //gtps. ddbj. nig. ac. jp/ n 2006/10/23 Updated once a year Primary Biological Databases 24

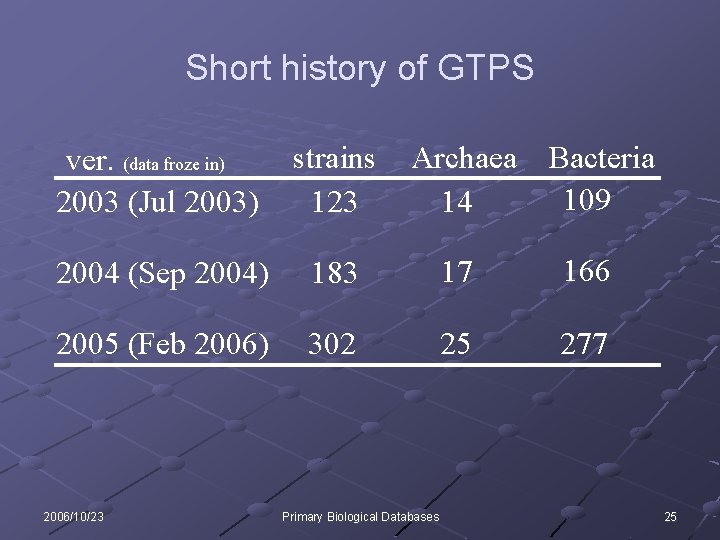
Short history of GTPS ver. (data froze in) 2003 (Jul 2003) strains 123 2004 (Sep 2004) 183 17 166 2005 (Feb 2006) 302 25 277 2006/10/23 Archaea Bacteria 109 14 Primary Biological Databases 25

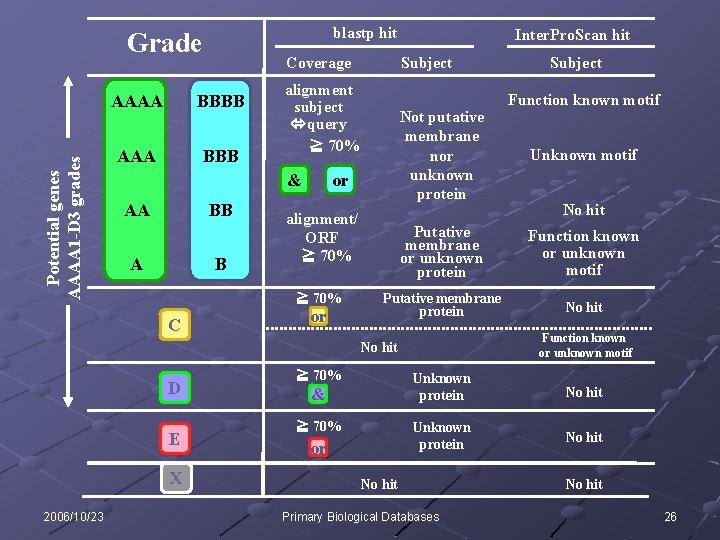
blastp hit Potential genes AAAA 1 -D 3 grades Grade Coverage AAAA BBBB AAA BBB AA BB A B Not putative membrane nor unknown protein or alignment/ ORF ≧ 70% C Subject alignment subject query ≧ 70% & or Inter. Pro. Scan hit E X 2006/10/23 Function known motif Unknown motif No hit Putative membrane or unknown protein Function known or unknown motif Putative membrane protein No hit Function known or unknown motif No hit D Subject ≧ 70% & ≧ 70% or Unknown protein No hit Primary Biological Databases No hit 26

Number of ORFs sorted by each grade AAAA-A GTPS ver. 2003 283, 247 GTPS ver. 2004 431, 672 GTPS ver. 2005 752, 186 BBBB-B 7, 208 10, 250 20, 755 C 4, 680 7, 511 16, 227 D 79, 779 107, 382 137, 656 E 6, 788 10, 225 17, 739 X 466, 681 687, 110 278, 075 Grades Total Potential genes (AAAA 1 - D 3) INSDC 2006/10/23 848, 383 1, 254, 150 Glimmer 2 and RBS finder 3 370, 876 362, 828 551, 246 537, 312 Primary Biological Databases 1, 222, 638 Glimmer 903, 845 904, 530 27

Comparison of the number of protein coding genes between GTPS and INSDC GTPS ver. 2003 GTPS ver. 2004 GTPS ver. 2005 Identical 261, 720 70. 6% 390, 557 70. 8% 576, 762 63. 8% 3' matched 92, 206 24. 9% 133, 146 24. 2% 252, 365 27. 9% New ORFs (Not annotated in INSDC) 14, 954 4. 0% 23, 935 4. 3% 31, 438 3. 5% Not predicted by Glimmer 1, 996 0. 5% 3, 608 0. 7% 43, 280 4. 8% Total (potential genes) 370, 876 551, 246 903, 845 Glimmer 3 was used for ver. 2005. 2006/10/23 Primary Biological Databases 28

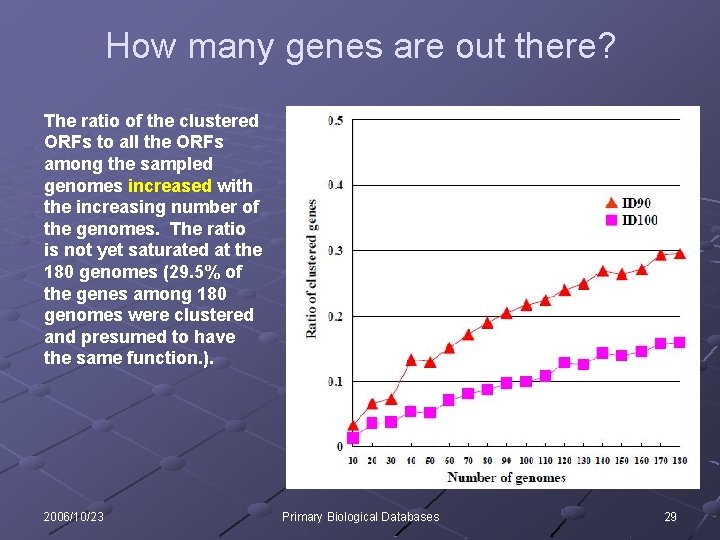
How many genes are out there? The ratio of the clustered ORFs to all the ORFs among the sampled genomes increased with the increasing number of the genomes. The ratio is not yet saturated at the 180 genomes (29. 5% of the genes among 180 genomes were clustered and presumed to have the same function. ). 2006/10/23 Primary Biological Databases 29

Summary It is the long term mission of the primary database to archive all the data published for the long term. At the same time, it is requested to provide objective/reliable data and tools for the mash-up. It proposes and maintains standards of biological data processing for the long term. 2006/10/23 Primary Biological Databases 30
 Nucleotide sequence vs amino acid sequence
Nucleotide sequence vs amino acid sequence International nucleotide sequence database collaboration
International nucleotide sequence database collaboration Dna vs rna
Dna vs rna Hgprt deficiency
Hgprt deficiency What type of bond connects the nitrogenous bases
What type of bond connects the nitrogenous bases Properties of nucleotide
Properties of nucleotide Pyramidine synthesis
Pyramidine synthesis Nucleotide metabolism
Nucleotide metabolism Nucleotide metabolism
Nucleotide metabolism Nucleotide nomenclature
Nucleotide nomenclature Pentose sugar structure in dna
Pentose sugar structure in dna Molecule of dna labeled
Molecule of dna labeled Phosphoanhydride bonds
Phosphoanhydride bonds Nucleotide to amino acid
Nucleotide to amino acid Nucleotide subunits
Nucleotide subunits Purine nucleotide cycle
Purine nucleotide cycle Nucleotide nomenclature
Nucleotide nomenclature Bioflix activity dna replication lagging strand synthesis
Bioflix activity dna replication lagging strand synthesis Structure name
Structure name Nucleotide in a sentence
Nucleotide in a sentence Nucleotide consists of
Nucleotide consists of Nucleotide nitrogenous base
Nucleotide nitrogenous base Store and transmit hereditary
Store and transmit hereditary Nucleotide acid
Nucleotide acid Dna nucleotide
Dna nucleotide Cyrasinc
Cyrasinc Deoxyribose nucleotide
Deoxyribose nucleotide Nucleotide nitrogenous base
Nucleotide nitrogenous base Dna nucleotide
Dna nucleotide Single nucleotide polymorphism (snp)
Single nucleotide polymorphism (snp) What is a nucleotide
What is a nucleotide