PYTHON WHAT IS BIOPYTHON Biopython is a python

PYTHON
WHAT IS BIOPYTHON? Biopython is a python library of resources for developers of Python-base software for bioinformatics and research. • can parse bioinformatics files into local data structures • Fasta, Gen. Bank, Blast output Clustalw etc. • Can access many files directly ( web database, NCBI) from within the script. • Works with sequences and records • Many search algorithms, comparative algorithms and format options.

INSTALLING BIOPYTHON Comes with Anaconda. You don’t even have to type in the import commands! If you use the standard IDLE environment you will need to download Bio. Python and place it in the proper directory. Bioinformatics has become so important in recent years that almost every programming environment, C++, Perl, etc has its own Bioinfo libraries.
SEQUENCE OBJECTS Biological sequences represent the main point of interest in Bioinformatics processing. Python includes a special datatype called a Sequence objects are not the same as Python strings. They are really strings together with additional information, such as an alphabet, and a variety of methods such as translate(), reverse_complement() and so on. dna = ‘AGTACACTGGT ‘ this is a pure string // Here is how you create a sequence object. seqdna = Seq(‘AGTACACTGGT ‘, Alphabet()) sequence obj Note that seqdna is a sequence object not just a string.
ALPHABETS - SEE IUPAC (INTERNATIONAL UNION OF PURE AND APPLIED CHEMISTRY) Alphabets are just the set of allowable characters that are used in the string. IUPAC. unambiguous_dna is really just the set {A, C, G, T} of nucleotides. IUPAC. unambiguous_rna is {A, C, G, U} IUPAC. protein is just the 20 standard amino acids {A, R, N, D, C, Q, E, H, I, L, K, M, F, P, S, T, W, Y, V} and others We will use mainly the {A, C, G, T} DNA set. Nice for type checking our sequences.

DUMPING ALPHABETS from Bio. Alphabet import IUPAC print IUPAC. unambiguous_dna. letters print IUPAC. unambiguous_rna. letters print IUPAC. protein. letters OUTPUT GATCRYWSMKHBVDN GAUC ACDEFGHIKLMNPQRSTVWY
CAN WORK WITH SEQUENCE OBJECTS LIKE STRINGS from Bio. Seq import Seq from Bio. Alphabet import IUPAC my_seq = Seq("GATCG", IUPAC. unambiguous_dna) print my_seq[0] prints first letter print len(my_seq) print length of string in sequence print Seq(“AAAA”). count(“AA”) non overlapping count ie 2 print GC(my_seq) Gives the GC % of the sequence. print my_seq[2: 5] We can even slice them. Returns a Seq. #convert seq obj to a pure string obj dna_string = str(my_seq)

NUCLEOTIDE SEQUENCES AND (REVERSE) COMPLEMENTS >>> from Bio. Seq import Seq >>> from Bio. Alphabet import IUPAC >>> my_seq = Seq("GATCGATGGGCCTATATAGGATCGAAAATCGC", IUPAC. unambiguous_dna) >>> my_seq Seq('GATCGATGGGCCTATATAGGATCGAAAATCGC', IUPACUnambiguous. DNA()) >>> my_seq. complement() Seq('CTAGCTACCCGGATATATCCTAGCTTTTAGCG', IUPACUnambiguous. DNA()) >>> my_seq. reverse_complement() Seq('GCGATTTTCGATCCTATATAGGCCCATCGATC', IUPACUnambiguous. DNA())
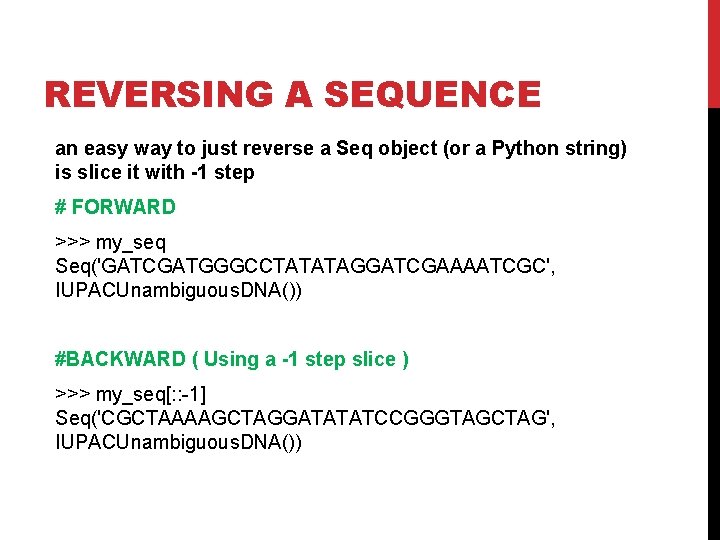
REVERSING A SEQUENCE an easy way to just reverse a Seq object (or a Python string) is slice it with -1 step # FORWARD >>> my_seq Seq('GATCGATGGGCCTATATAGGATCGAAAATCGC', IUPACUnambiguous. DNA()) #BACKWARD ( Using a -1 step slice ) >>> my_seq[: : -1] Seq('CGCTAAAAGCTAGGATATATCCGGGTAGCTAG', IUPACUnambiguous. DNA())

DOUBLE STRANDED DNA coding strand (aka Crick strand, strand +1) 5’ ATGGCCATTGTAATGGGCCGCTGAAAGGGTGCCCGATAG 3’ |||||||||||||||||||| 3’ TACCGGTAACATTACCCGGCGACTTTCCCACGGGCTATC 5’ DNA template strand (aka Watson strand, strand − 1)
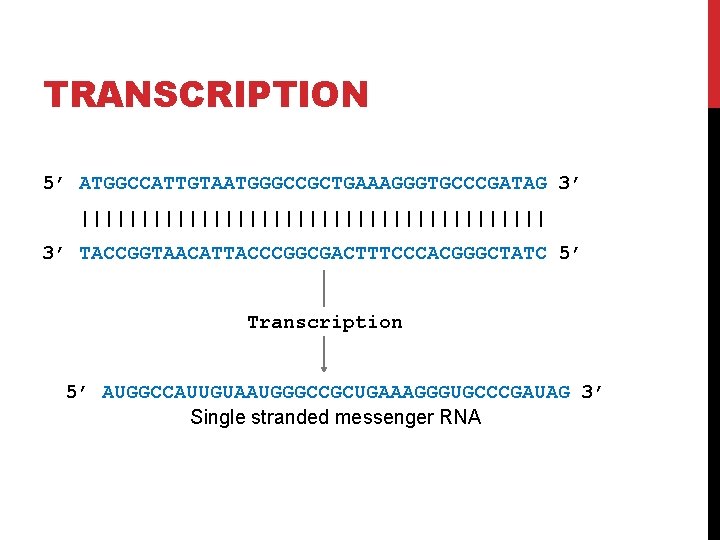
TRANSCRIPTION 5’ ATGGCCATTGTAATGGGCCGCTGAAAGGGTGCCCGATAG 3’ |||||||||||||||||||| 3’ TACCGGTAACATTACCCGGCGACTTTCCCACGGGCTATC 5’ Transcription 5’ AUGGCCAUUGUAAUGGGCCGCUGAAAGGGUGCCCGAUAG 3’ Single stranded messenger RNA

LETS DO SOME REVERSE COMP from Bio. Seq import Seq from Bio. Alphabet import IUPAC coding_dna = Seq(“ATGGCCATTGTAATGGGCCGCTGAAAGGGTGCCCGATAG", IUPAC. unambiguous_dna) template_dna= coding_dna. reverse_complement() print template_dna CTATCGGGCACCCTTTCAGCGGCCCATTACAATGGCCAT
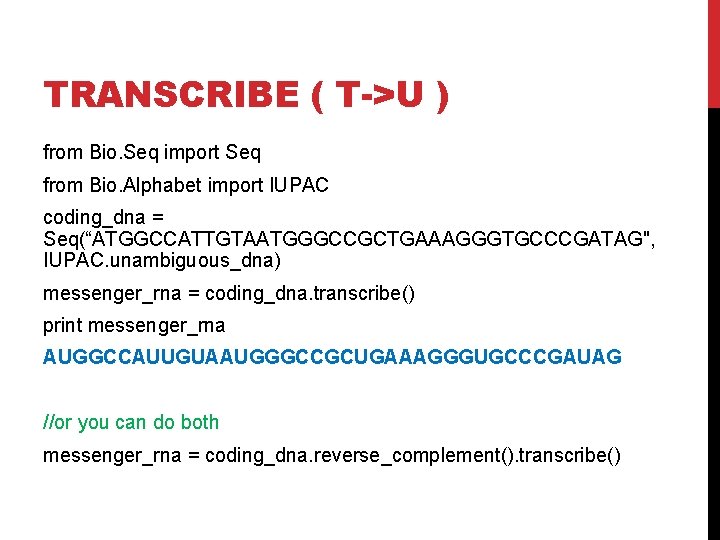
TRANSCRIBE ( T->U ) from Bio. Seq import Seq from Bio. Alphabet import IUPAC coding_dna = Seq(“ATGGCCATTGTAATGGGCCGCTGAAAGGGTGCCCGATAG", IUPAC. unambiguous_dna) messenger_rna = coding_dna. transcribe() print messenger_rna AUGGCCAUUGUAAUGGGCCGCUGAAAGGGUGCCCGAUAG //or you can do both messenger_rna = coding_dna. reverse_complement(). transcribe()

TRANSLATE INTO PROTEIN from Bio. Seq import Seq from Bio. Alphabet import IUPAC messenger_rna = Seq("AUGGCCAUUGUAAUGGGCCGCUGAAAGGGUGCCCG AUAG", IUPAC. unambiguous_rna) print messenger_rna. translate() # I added the spaces AUG GCC AUU GUA AUG GGC CGC UGA AAG GGU GCC CGA UAG MAIVMGR*KGAR* # the * represents stop codons.
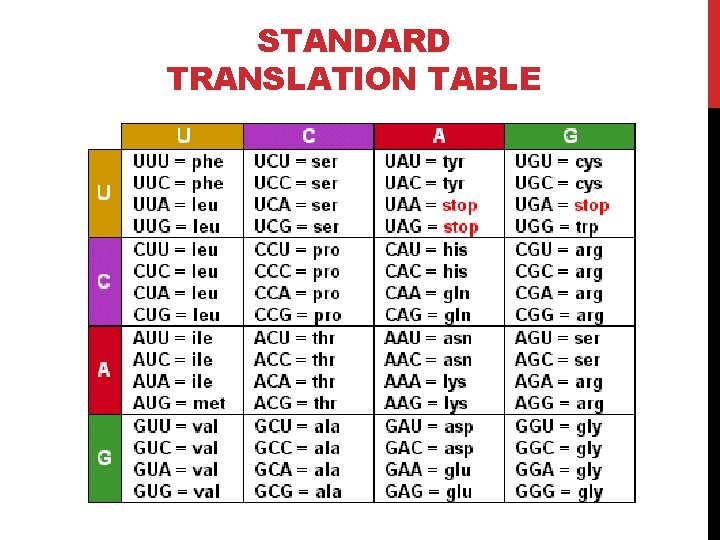
STANDARD TRANSLATION TABLE

PRINTING TABLES from Bio. Seq import Seq from Bio. Alphabet import IUPAC from Bio. Data import Codon. Table std. Table = Codon. Table. unambiguous_dna_by_id[1] print std. Table mito. Table = Codon. Table. unambiguous_dna_by_id[2] print mito. Table
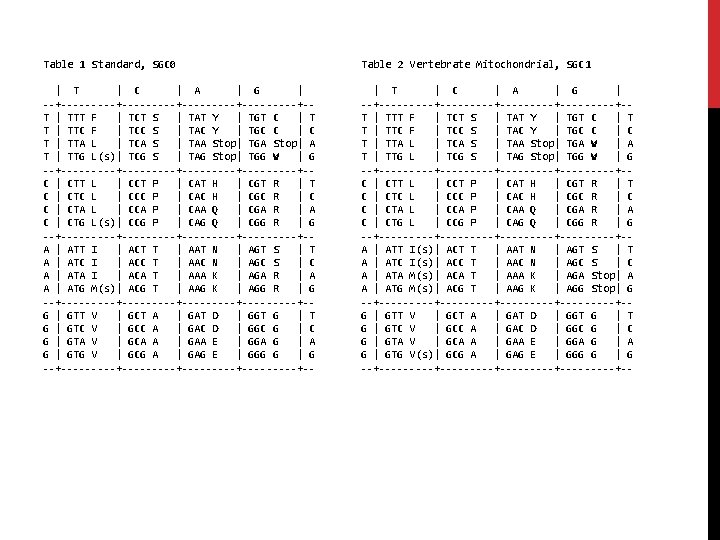
Table 1 Standard, SGC 0 Table 2 Vertebrate Mitochondrial, SGC 1 | T | C | A | G | --+---------+---------+-T | TTT F | TCT S | TAT Y | TGT C | T T | TTC F | TCC S | TAC Y | TGC C | C T | TTA L | TCA S | TAA Stop| TGA Stop| A T | TTG L(s)| TCG S | TAG Stop| TGG W | G --+---------+---------+-C | CTT L | CCT P | CAT H | CGT R | T C | CTC L | CCC P | CAC H | CGC R | C C | CTA L | CCA P | CAA Q | CGA R | A C | CTG L(s)| CCG P | CAG Q | CGG R | G --+---------+---------+-A | ATT I | ACT T | AAT N | AGT S | T A | ATC I | ACC T | AAC N | AGC S | C A | ATA I | ACA T | AAA K | AGA R | A A | ATG M(s)| ACG T | AAG K | AGG R | G --+---------+---------+-G | GTT V | GCT A | GAT D | GGT G | GTC V | GCC A | GAC D | GGC G | GTA V | GCA A | GAA E | GGA G | GTG V | GCG A | GAG E | GGG G | G --+---------+---------+-- | T | C | A | G | --+---------+---------+-T | TTT F | TCT S | TAT Y | TGT C | T T | TTC F | TCC S | TAC Y | TGC C | C T | TTA L | TCA S | TAA Stop| TGA W | A T | TTG L | TCG S | TAG Stop| TGG W | G --+---------+---------+-C | CTT L | CCT P | CAT H | CGT R | T C | CTC L | CCC P | CAC H | CGC R | C C | CTA L | CCA P | CAA Q | CGA R | A C | CTG L | CCG P | CAG Q | CGG R | G --+---------+---------+-A | ATT I(s)| ACT T | AAT N | AGT S | T A | ATC I(s)| ACC T | AAC N | AGC S | C A | ATA M(s)| ACA T | AAA K | AGA Stop| A A | ATG M(s)| ACG T | AAG K | AGG Stop| G --+---------+---------+-G | GTT V | GCT A | GAT D | GGT G | GTC V | GCC A | GAC D | GGC G | GTA V | GCA A | GAA E | GGA G | GTG V(s)| GCG A | GAG E | GGG G | G --+---------+---------+--
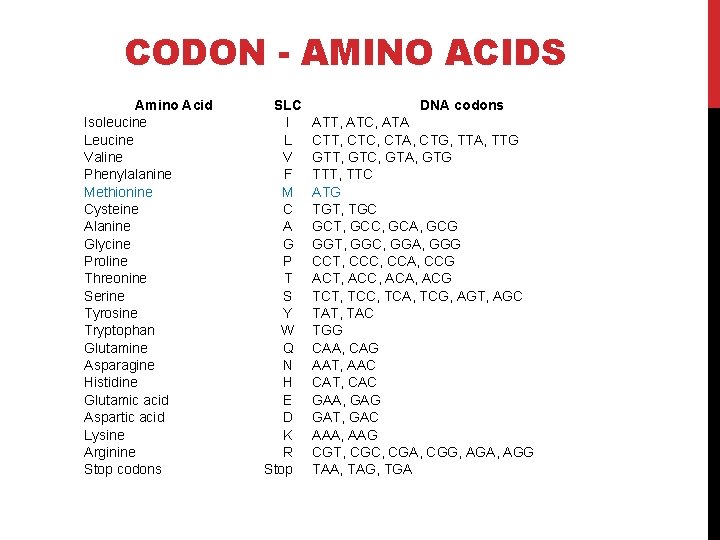
CODON - AMINO ACIDS Amino Acid Isoleucine Leucine Valine Phenylalanine Methionine Cysteine Alanine Glycine Proline Threonine Serine Tyrosine Tryptophan Glutamine Asparagine Histidine Glutamic acid Aspartic acid Lysine Arginine Stop codons . SLC I L V F M C A G P T S Y W Q N H E D K R Stop DNA codons ATT, ATC, ATA CTT, CTC, CTA, CTG, TTA, TTG GTT, GTC, GTA, GTG TTT, TTC ATG TGT, TGC GCT, GCC, GCA, GCG GGT, GGC, GGA, GGG CCT, CCC, CCA, CCG ACT, ACC, ACA, ACG TCT, TCC, TCA, TCG, AGT, AGC TAT, TAC TGG CAA, CAG AAT, AAC CAT, CAC GAA, GAG GAT, GAC AAA, AAG CGT, CGC, CGA, CGG, AGA, AGG TAA, TAG, TGA

THE SEQRECORD OBJECT A Seq. Record is a structure that allows the storage of additional information with a sequence. This includes the usual information found in standard genbank files. The following is a sample. . seq - The sequence. id - The primary ID used to identify the sequence (String). name – The common name of the sequence. annotations – A dictionary of additional information about the sequence. features –A list of Seq. Feature objects

READ A RECORD from Bio import Seq. IO record = Seq. IO. read("micoplasma. Gen. gb", "genbank") print record. description ct=0 for f in record. features: if f. type=='gene': ct+=1 print ct
- Slides: 20