Putting the Pieces Together Integrating Computational Tools to



- Slides: 20

Putting the Pieces Together: Integrating Computational Tools to Characterize Protein Expression in Bio. Energy-Related Organisms Rachel Adams, Richard Giannone, Paul Abraham, Robert Hettich

Emphasis on Science Integration Better Plants Better Microbes Better Tools and Combinations 2

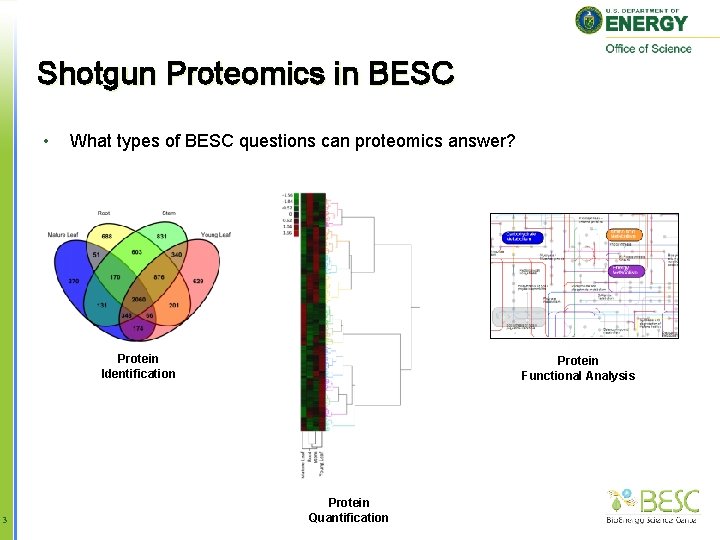
Shotgun Proteomics in BESC • What types of BESC questions can proteomics answer? Protein Identification 3 Protein Functional Analysis Protein Quantification

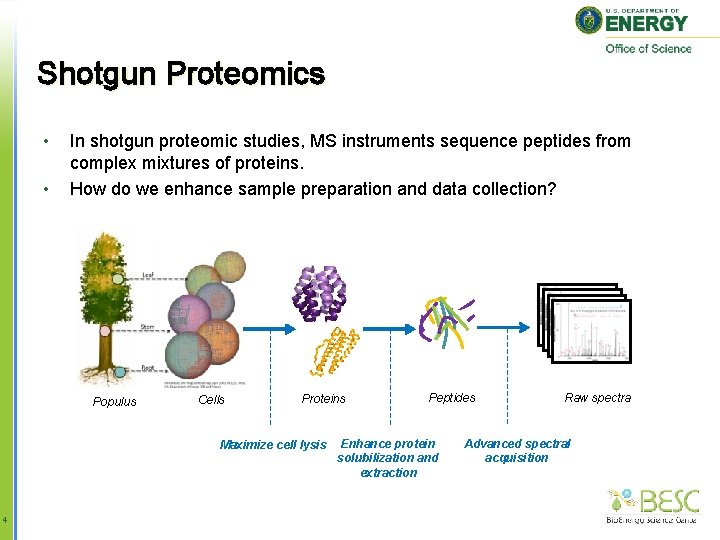
Shotgun Proteomics • • In shotgun proteomic studies, MS instruments sequence peptides from complex mixtures of proteins. How do we enhance sample preparation and data collection? Populus Cells Proteins Maximize cell lysis 4 Peptides Enhance protein solubilization and extraction Raw spectra Advanced spectral acquisition

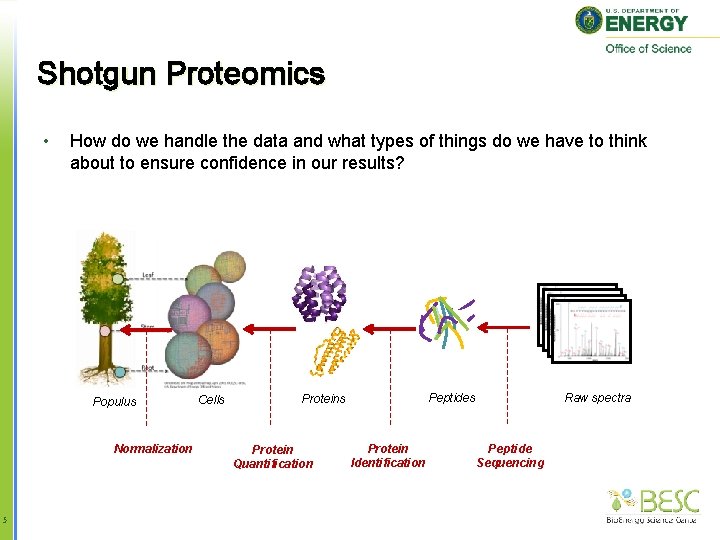
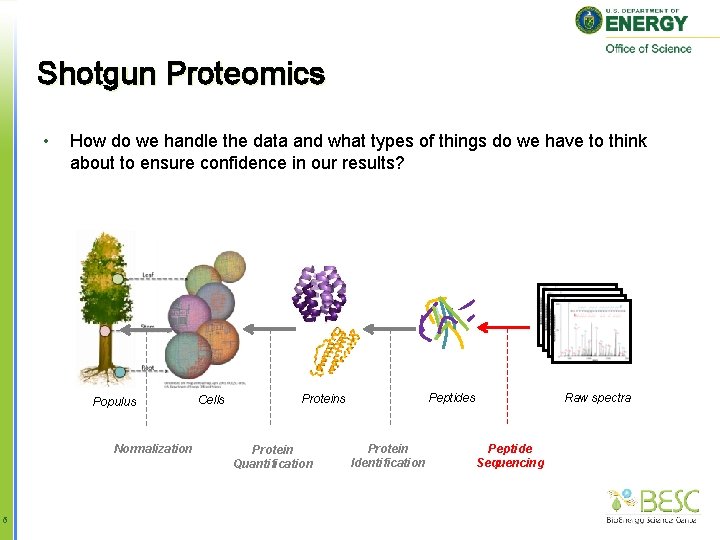
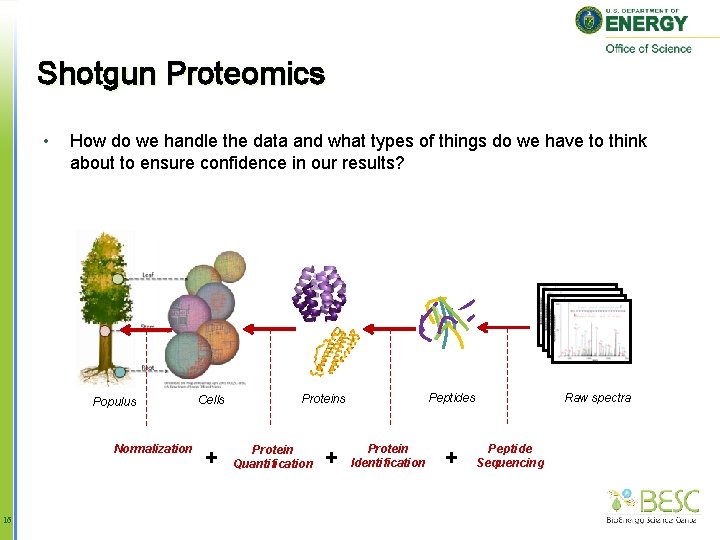
Shotgun Proteomics • How do we handle the data and what types of things do we have to think about to ensure confidence in our results? Populus Normalization 5 Cells Protein Quantification Raw spectra Peptides Protein Identification Peptide Sequencing

Shotgun Proteomics • How do we handle the data and what types of things do we have to think about to ensure confidence in our results? Populus Normalization 6 Cells Protein Quantification Raw spectra Peptides Protein Identification Peptide Sequencing

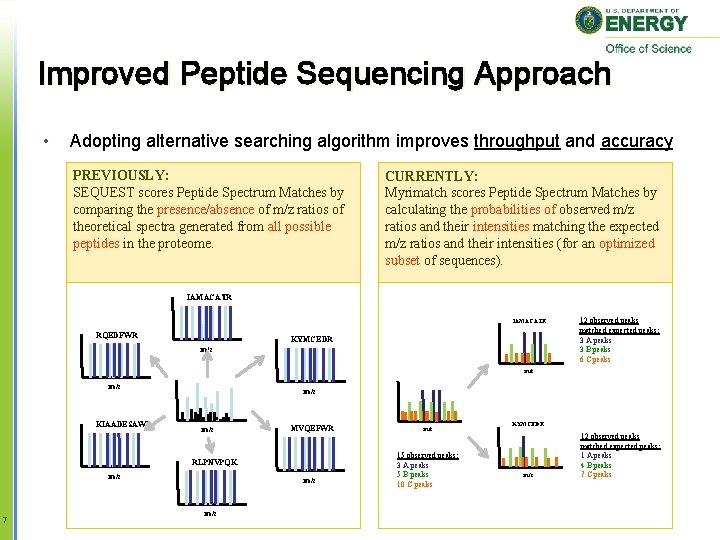
Improved Peptide Sequencing Approach • Adopting alternative searching algorithm improves throughput and accuracy PREVIOUSLY: SEQUEST scores Peptide Spectrum Matches by comparing the presence/absence of m/z ratios of theoretical spectra generated from all possible peptides in the proteome. CURRENTLY: Myrimatch scores Peptide Spectrum Matches by calculating the probabilities of observed m/z ratios and their intensities matching the expected m/z ratios and their intensities (for an optimized subset of sequences). IAMACATR RQEDFWR KYMCEDR m/z 12 observed peaks matched expected peaks: 3 A peaks 3 B peaks 6 C peaks m/z KIAADESAWR m/z MVQEFWR RLPNVPQK m/z 7 m/z m/z 15 observed peaks: 3 A peaks 5 B peaks 10 C peaks KYMCEDR m/z 12 observed peaks matched expected peaks: 1 A peaks 4 B peaks 7 C peaks

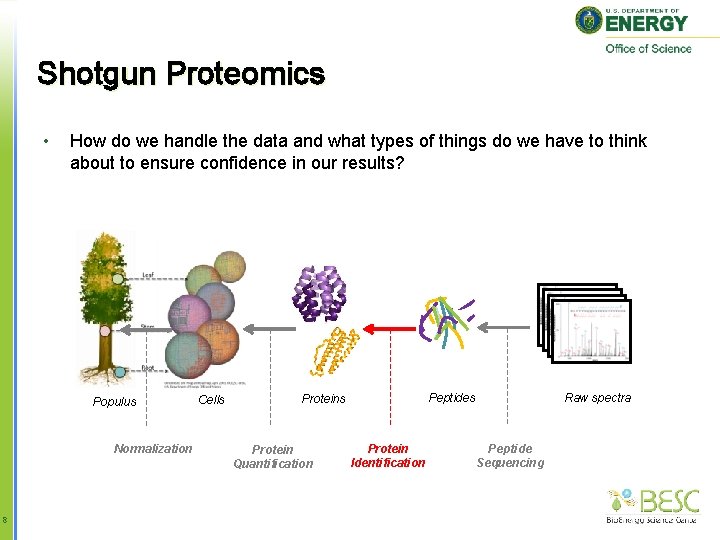
Shotgun Proteomics • How do we handle the data and what types of things do we have to think about to ensure confidence in our results? Populus Normalization 8 Cells Protein Quantification Raw spectra Peptides Protein Identification Peptide Sequencing

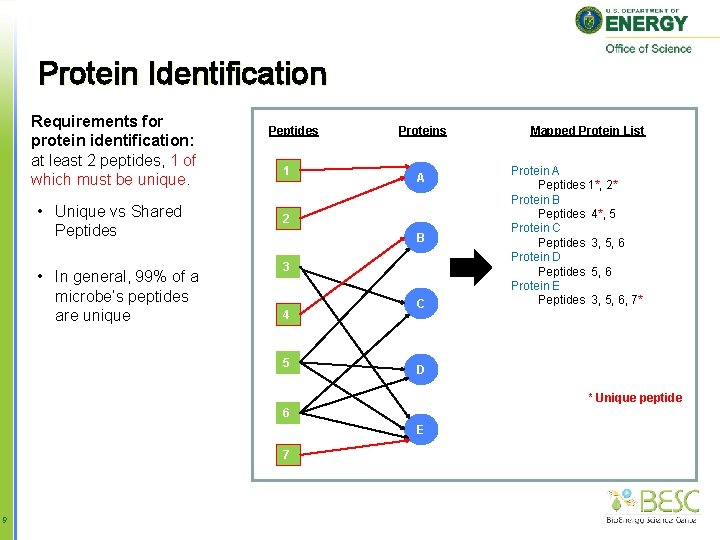
Protein Identification Requirements for protein identification: at least 2 peptides, 1 of which must be unique. • Unique vs Shared Peptides • In general, 99% of a microbe’s peptides are unique Peptides 1 Proteins A 2 B 3 4 5 C Mapped Protein List Protein A Peptides 1*, 2* Protein B Peptides 4*, 5 Protein C Peptides 3, 5, 6 Protein D Peptides 5, 6 Protein E Peptides 3, 5, 6, 7* D * Unique peptide 6 E 7 9

Protein Redundancy in Plants • 10 Populus has a complex genome… which translates into a complex proteome – Genome version 2. 2 has 45, 778 genes – 10% of the gene models >95% similarity – 309 instances of tandem repeats – 140 genes with > 2 alternative splice variants

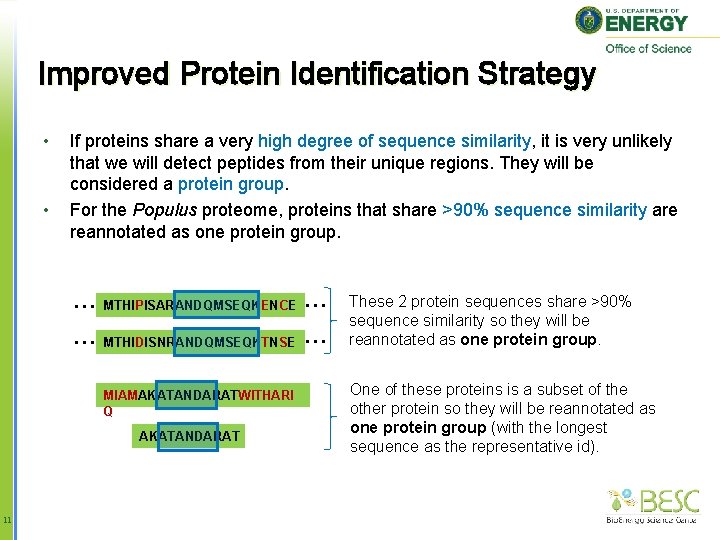
Improved Protein Identification Strategy • • If proteins share a very high degree of sequence similarity, it is very unlikely that we will detect peptides from their unique regions. They will be considered a protein group. For the Populus proteome, proteins that share >90% sequence similarity are reannotated as one protein group. … MTHIPISARANDQMSEQKENCE … … MTHIDISNRANDQMSEQKTNSE … MIAMAKATANDARATWITHARI Q AKATANDARAT 11 These 2 protein sequences share >90% sequence similarity so they will be reannotated as one protein group. One of these proteins is a subset of the other protein so they will be reannotated as one protein group (with the longest sequence as the representative id).

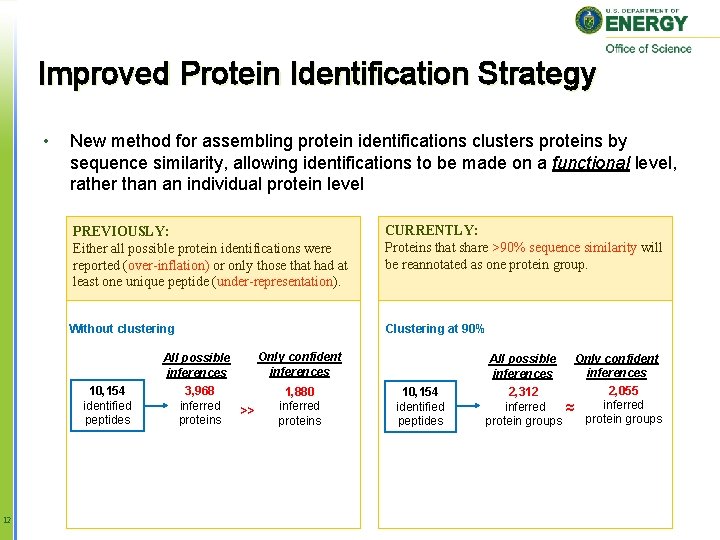
Improved Protein Identification Strategy • New method for assembling protein identifications clusters proteins by sequence similarity, allowing identifications to be made on a functional level, rather than an individual protein level PREVIOUSLY: Either all possible protein identifications were reported (over-inflation) or only those that had at least one unique peptide (under-representation). CURRENTLY: Proteins that share >90% sequence similarity will be reannotated as one protein group. Without clustering Clustering at 90% Only confident inferences All possible inferences 10, 154 identified peptides 12 3, 968 inferred proteins >> 1, 880 inferred proteins Only confident inferences 2, 055 2, 312 inferred ≈ protein groups All possible inferences 10, 154 identified peptides

Shotgun Proteomics • How do we handle the data and what types of things do we have to think about to ensure confidence in our results? Populus Normalization 13 Cells Protein Quantification Raw spectra Peptides Protein Identification Peptide Sequencing

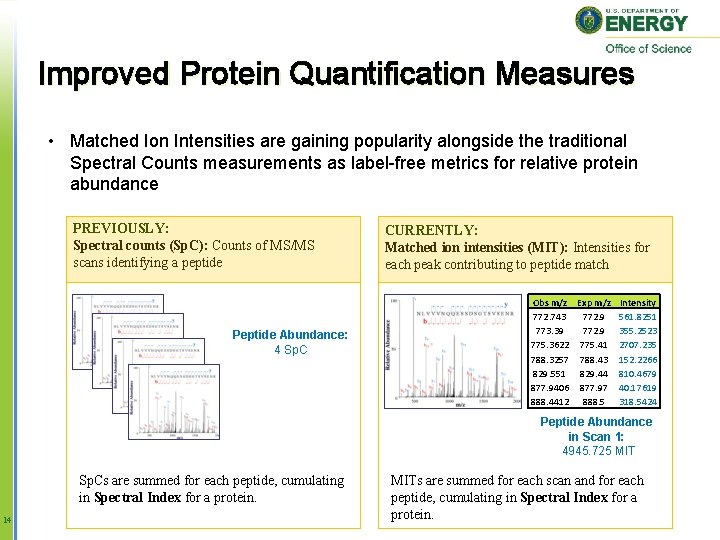
Improved Protein Quantification Measures • Matched Ion Intensities are gaining popularity alongside the traditional Spectral Counts measurements as label-free metrics for relative protein abundance PREVIOUSLY: Spectral counts (Sp. C): Counts of MS/MS scans identifying a peptide Peptide Abundance: 4 Sp. C CURRENTLY: Matched ion intensities (MIT): Intensities for each peak contributing to peptide match Obs m/z Exp m/z 772. 743 772. 9 773. 39 772. 9 775. 3622 775. 41 788. 3257 788. 43 829. 551 829. 44 877. 9406 877. 97 888. 4412 888. 5 Intensity 561. 8251 355. 2523 2707. 235 152. 2266 810. 4679 40. 17619 318. 5424 Peptide Abundance in Scan 1: 4945. 725 MIT Sp. Cs are summed for each peptide, cumulating in Spectral Index for a protein. 14 MITs are summed for each scan and for each peptide, cumulating in Spectral Index for a protein.

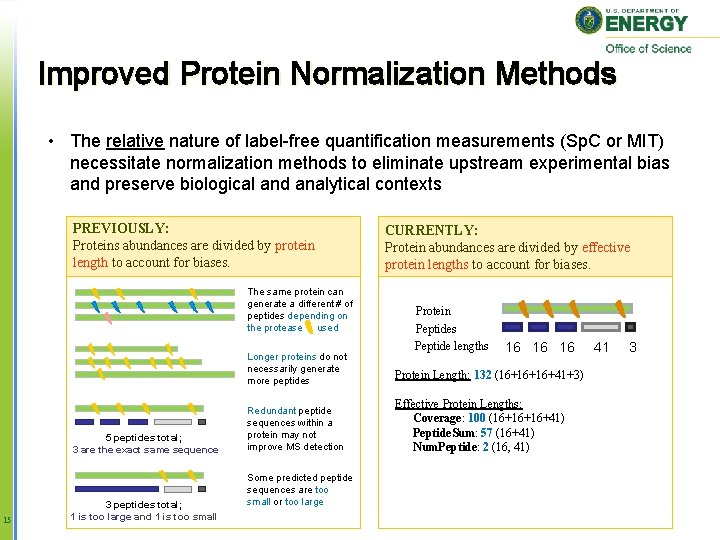
Improved Protein Normalization Methods • The relative nature of label-free quantification measurements (Sp. C or MIT) necessitate normalization methods to eliminate upstream experimental bias and preserve biological and analytical contexts PREVIOUSLY: Proteins abundances are divided by protein length to account for biases. The same protein can generate a different # of peptides depending on the protease used 5 peptides total; 3 are the exact same sequence 15 3 peptides total; 1 is too large and 1 is too small CURRENTLY: Protein abundances are divided by effective protein lengths to account for biases. Protein Peptides Peptide lengths 16 16 16 Longer proteins do not necessarily generate more peptides Protein Length: 132 (16+16+16+41+3) Redundant peptide sequences within a protein may not improve MS detection Effective Protein Lengths: Coverage: 100 (16+16+16+41) Peptide. Sum: 57 (16+41) Num. Peptide: 2 (16, 41) Some predicted peptide sequences are too small or too large 41 3

Shotgun Proteomics • How do we handle the data and what types of things do we have to think about to ensure confidence in our results? Populus Normalization 16 Cells + Protein Quantification + Raw spectra Peptides Protein Identification + Peptide Sequencing

PUTTING THE PIECES TOGETHER Cyber-infrastructure integrates hardware and software solutions to address growing demands of complex biological datasets 17

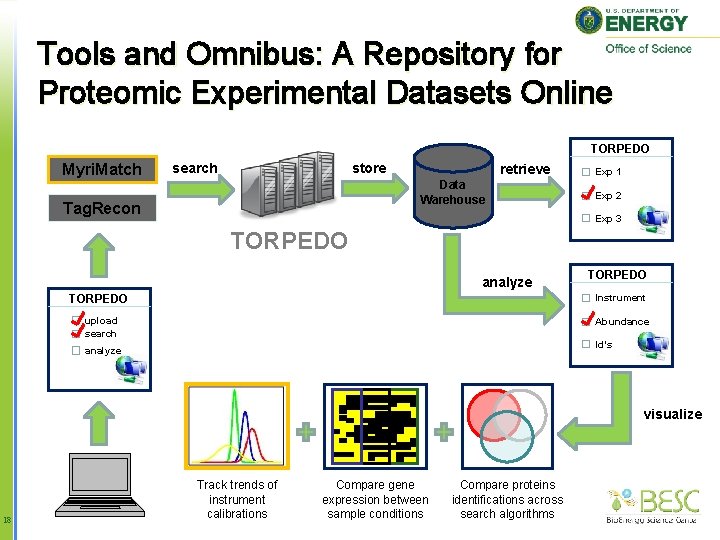
Tools and Omnibus: A Repository for Proteomic Experimental Datasets Online TORPEDO Myri. Match search store retrieve Data Warehouse Tag. Recon Exp 1 Exp 2 Exp 3 TORPEDO analyze TORPEDO Instrument upload search Abundance Id’s analyze visualize 18 Track trends of instrument calibrations Compare gene expression between sample conditions Compare proteins identifications across search algorithms

Conclusions l While it is important for informatic tools to be able to handle large datasets, it is becoming increasingly crucial for tools to also handle the biological complexity associated with more intricate experimental designs. Although some existing tools can scale computationally and maintain biological relevance, most of the time new tools need to be developed to appropriately address these concerns. l The overwhelming volume and complexity of these experiments requires that the new and existing tools are not only optimized for speed and interpretation, but they also necessitate seamless communication with each other in an integrated workflow. l By constructing a workflow that allows high-throughput processing of massive datasets, data collected within the past decade can be standardized and updated with the most recent analyses. Once these analyses are complete, meta-analyses can identify global analytical and biological trends. 19

Acknowledgements BESC Proteomics Robert Hettich Rich Giannone Paul Abraham Trish Lankford Weili Xiong Adam Martin BESC Plant Sciences Jerry Tuskan Udaya Kalluri Oak Ridge National Laboratory/DOE Organic and Biological Mass Spectrometry Group Chemical Sciences Division Biosciences Division Bio. Energy Science Center Advanced Scientific Computing Research Genome Science and Technology SCALE-IT NSF IGERT Fellowship Journal cover of Molecular & Cellular Proteomics, January 2013. Abraham et al. 20