Purdue University Cytometry Laboratories Functional assays Principles and
Purdue University Cytometry Laboratories Functional assays Principles and Methods J Paul Robinson These slides are on the PURDUE CYTOMETRY WEB SITE http: //flowcyt. cyto. purdue. edu Presented at the Polish Society for Cytometry Meeting, Gdansk, Poland, October 18, 1998
Poland taken from Comsat C 1 satellite October 15, 1998 you can clearly see Warsaw in the center. Top centerleft is Gdansk.

Gdansk Warsaw Krakow
Gdansk

The goals of this presentation are: • To identify the nature of functional assays in Cytometry • To expand on how they operate • To discuss the advantages and disadvantages of each • To discuss the application of these assays
Kinetics Principle of Time Measurements • Live cells can be measured as easily as dead cells • You only need a small number of cells in a changing environment • End point assays can describe the activity of the cell
Cellular Functions • Cell Viability • Phagocytosis • Organelle Function – mitochondria, ER – endosomes, Golgi • Oxidative Reactions – – Superoxide Hydrogen Peroxide Nitric Oxide Glutathione levels • Ionic Flux Determinations –Calcium –Intracellular p. H • Membrane Potential • Membrane Polarization • Lipid Peroxidation
Fluorescence What do we measure? TIME
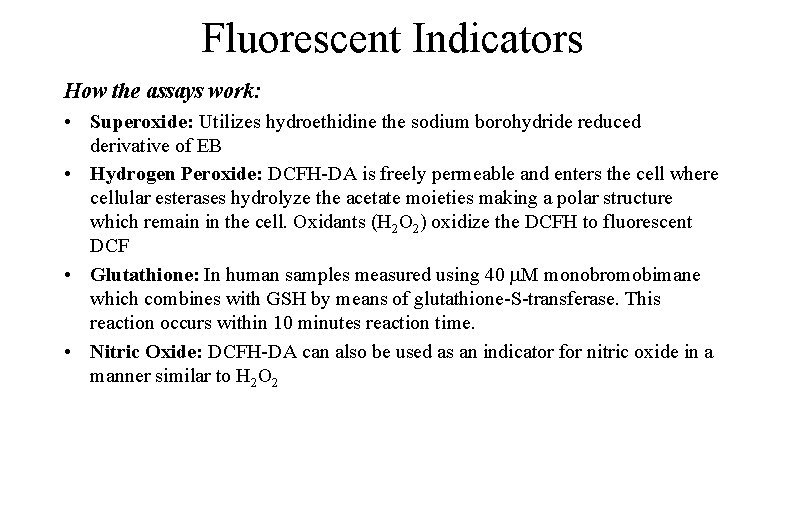
Fluorescent Indicators How the assays work: • Superoxide: Utilizes hydroethidine the sodium borohydride reduced derivative of EB • Hydrogen Peroxide: DCFH-DA is freely permeable and enters the cell where cellular esterases hydrolyze the acetate moieties making a polar structure which remain in the cell. Oxidants (H 2 O 2) oxidize the DCFH to fluorescent DCF • Glutathione: In human samples measured using 40 M monobromobimane which combines with GSH by means of glutathione-S-transferase. This reaction occurs within 10 minutes reaction time. • Nitric Oxide: DCFH-DA can also be used as an indicator for nitric oxide in a manner similar to H 2 O 2
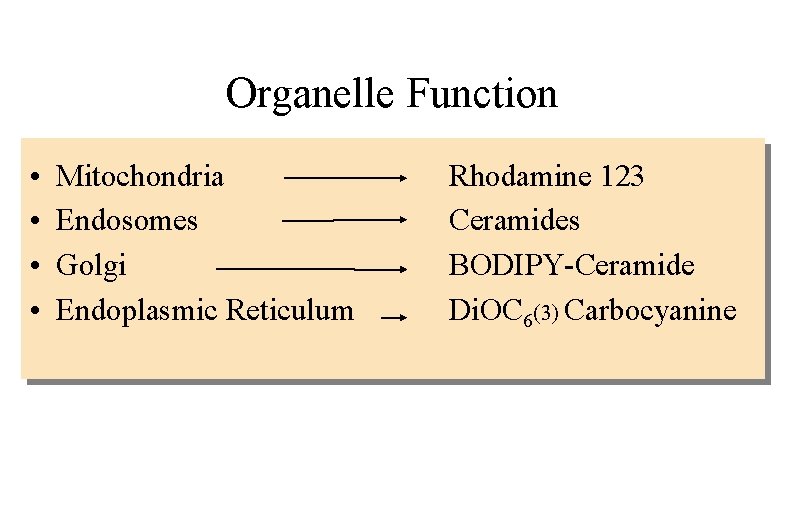
Organelle Function • • Mitochondria Endosomes Golgi Endoplasmic Reticulum Rhodamine 123 Ceramides BODIPY-Ceramide Di. OC 6(3) Carbocyanine
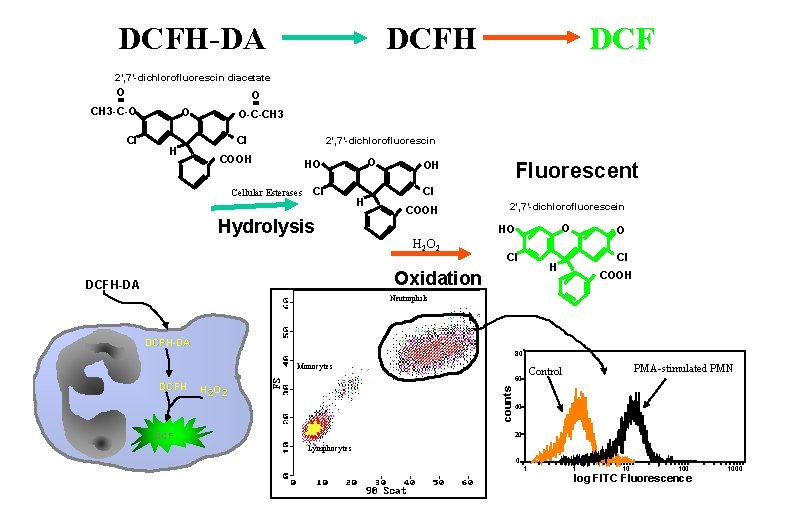
DCFH-DA DCFH DCF 2’, 7’-dichlorofluorescin diacetate O O CH 3 -C-O Cl O H O-C-CH 3 Cl 2’, 7’-dichlorofluorescin COOH O HO Cellular Esterases Cl Hydrolysis H Fluorescent OH Cl COOH 2’, 7’-dichlorofluorescein O HO H 2 O 2 Cl Cl H Oxidation DCFH-DA O COOH Neutrophils DCFH-DA 80 Monocytes H 2 O 2 counts DCFH 60 DCF PMA-stimulated PMN Control 40 20 Lymphocytes 0. 1 1 10 100 log FITC Fluorescence 1000
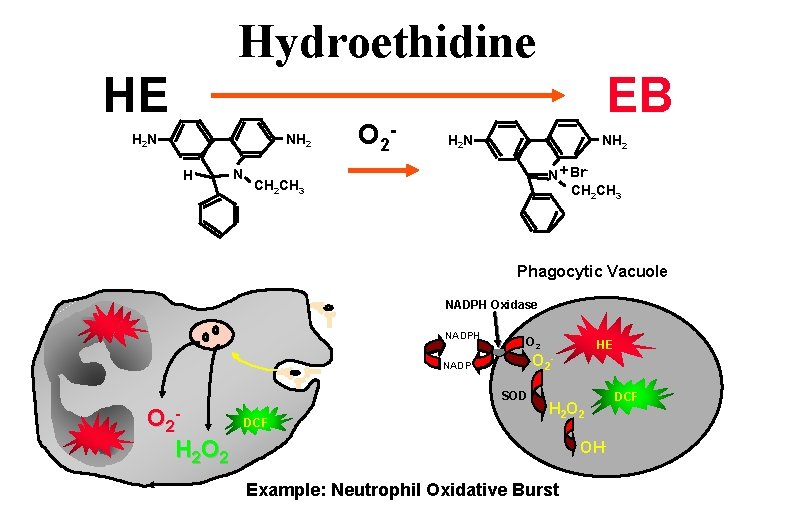
Hydroethidine HE H 2 N NH 2 H N O 2 - EB H 2 N NH 2 N + Br CH 2 CH 3 - CH 2 CH 3 Phagocytic Vacuole NADPH Oxidase NADPH O 2 NADP SOD O 2 H 2 O 2 DCF HE - H 2 OH- Example: Neutrophil Oxidative Burst DCF
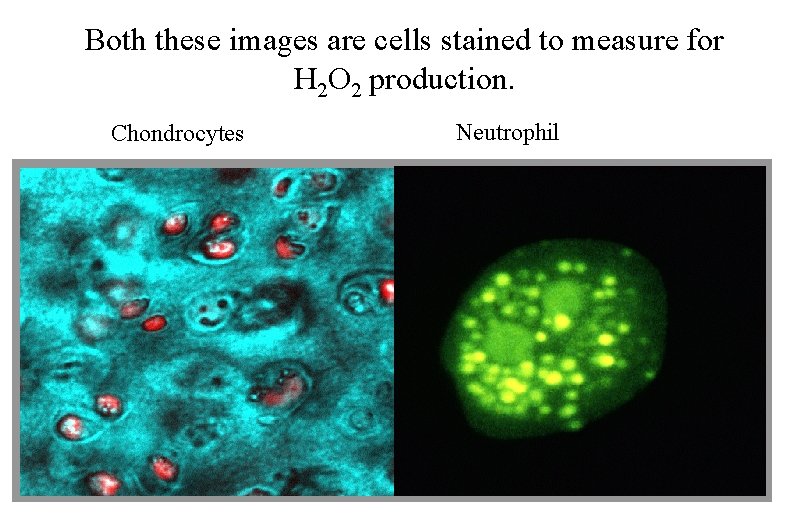
Both these images are cells stained to measure for H 2 O 2 production. Chondrocytes Neutrophil
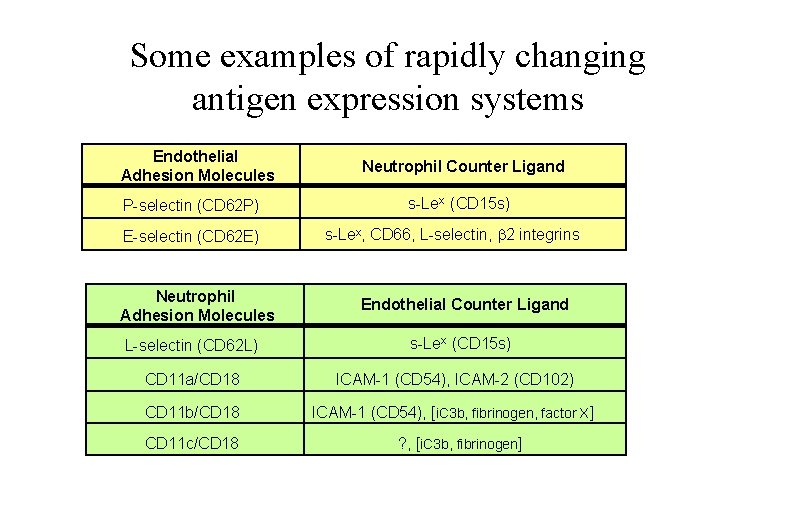
Some examples of rapidly changing antigen expression systems Endothelial Adhesion Molecules P-selectin (CD 62 P) E-selectin (CD 62 E) Neutrophil Adhesion Molecules L-selectin (CD 62 L) Neutrophil Counter Ligand s-Lex (CD 15 s) s-Lex, CD 66, L-selectin, b 2 integrins Endothelial Counter Ligand s-Lex (CD 15 s) CD 11 a/CD 18 ICAM-1 (CD 54), ICAM-2 (CD 102) CD 11 b/CD 18 ICAM-1 (CD 54), [i. C 3 b, fibrinogen, factor X] CD 11 c/CD 18 ? , [i. C 3 b, fibrinogen]
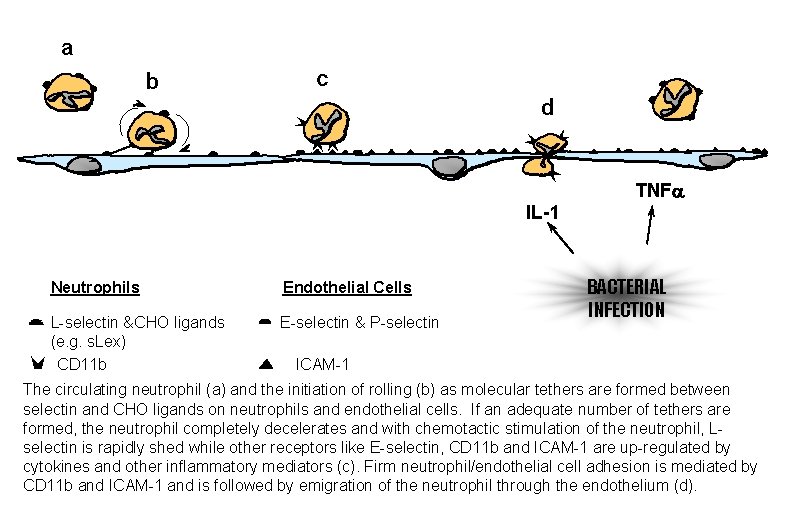
a b c d TNFa IL-1 Neutrophils Endothelial Cells L-selectin &CHO ligands (e. g. s. Lex) CD 11 b E-selectin & P-selectin BACTERIAL INFECTION ICAM-1 The circulating neutrophil (a) and the initiation of rolling (b) as molecular tethers are formed between selectin and CHO ligands on neutrophils and endothelial cells. If an adequate number of tethers are formed, the neutrophil completely decelerates and with chemotactic stimulation of the neutrophil, Lselectin is rapidly shed while other receptors like E-selectin, CD 11 b and ICAM-1 are up-regulated by cytokines and other inflammatory mediators (c). Firm neutrophil/endothelial cell adhesion is mediated by CD 11 b and ICAM-1 and is followed by emigration of the neutrophil through the endothelium (d).
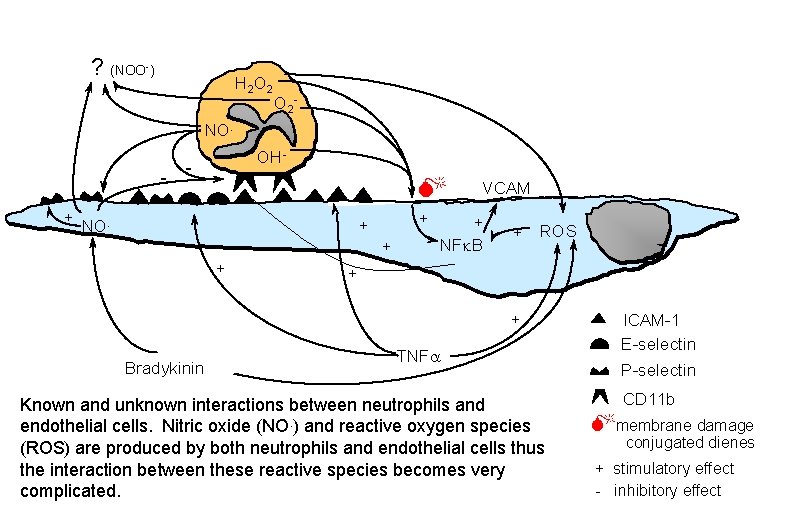
? (NOO-) H 2 O 2 - NO. - OH- - M + NO. + + VCAM + NFk. B + ROS + + Bradykinin TNFa Known and unknown interactions between neutrophils and endothelial cells. Nitric oxide (NO. ) and reactive oxygen species (ROS) are produced by both neutrophils and endothelial cells thus the interaction between these reactive species becomes very complicated. ICAM-1 E-selectin P-selectin CD 11 b Mmembrane damage conjugated dienes + stimulatory effect - inhibitory effect
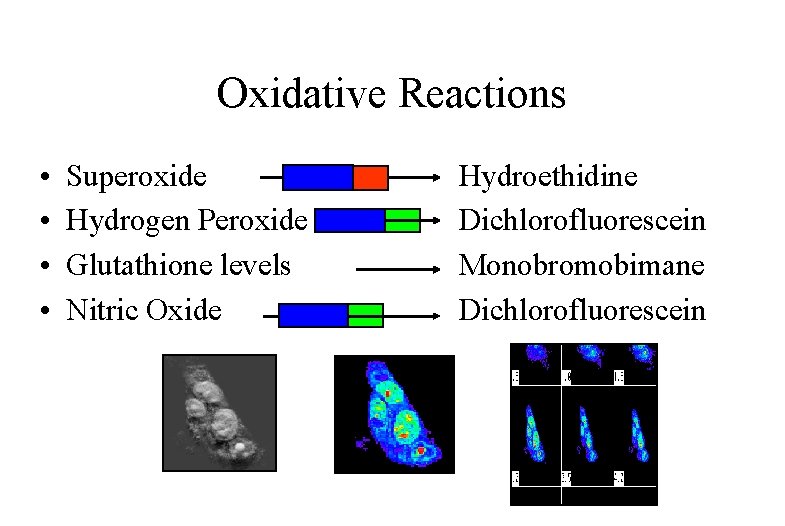
Oxidative Reactions • • Superoxide Hydrogen Peroxide Glutathione levels Nitric Oxide Hydroethidine Dichlorofluorescein Monobromobimane Dichlorofluorescein
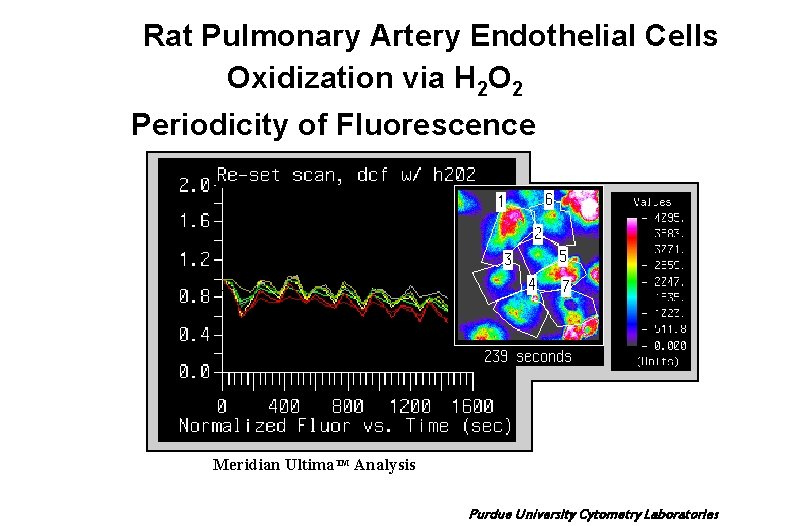
Rat Pulmonary Artery Endothelial Cells Oxidization via H 2 O 2 Periodicity of Fluorescence Meridian Ultima. TM Analysis Purdue University Cytometry Laboratories
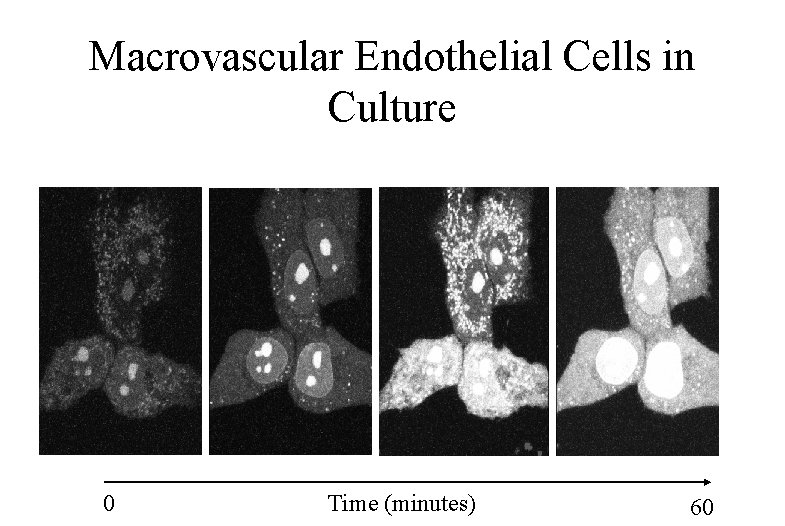
Macrovascular Endothelial Cells in Culture 0 Time (minutes) 60
Confocal System Culture System
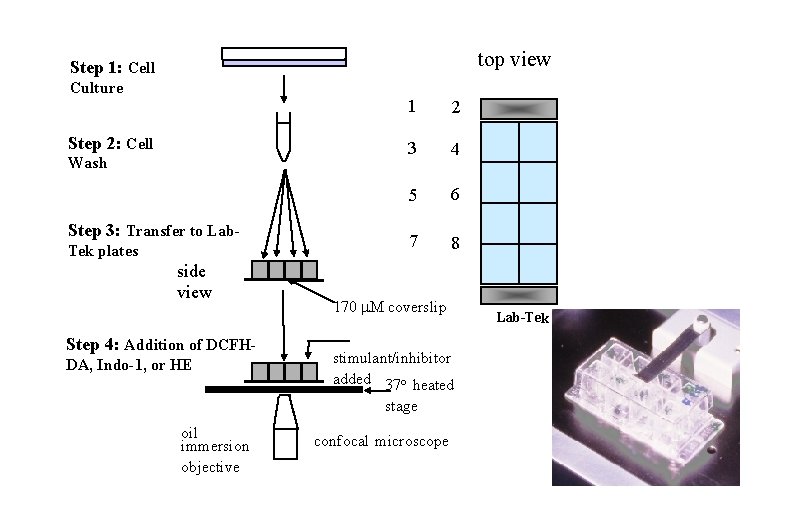
top view Step 1: Cell Culture Step 2: Cell Wash Step 3: Transfer to Lab. Tek plates side view Step 4: Addition of DCFHDA, Indo-1, or HE 1 2 3 4 5 6 7 8 170 M coverslip stimulant/inhibitor added 37 o heated stage oil immersion objective confocal microscope Lab-Tek
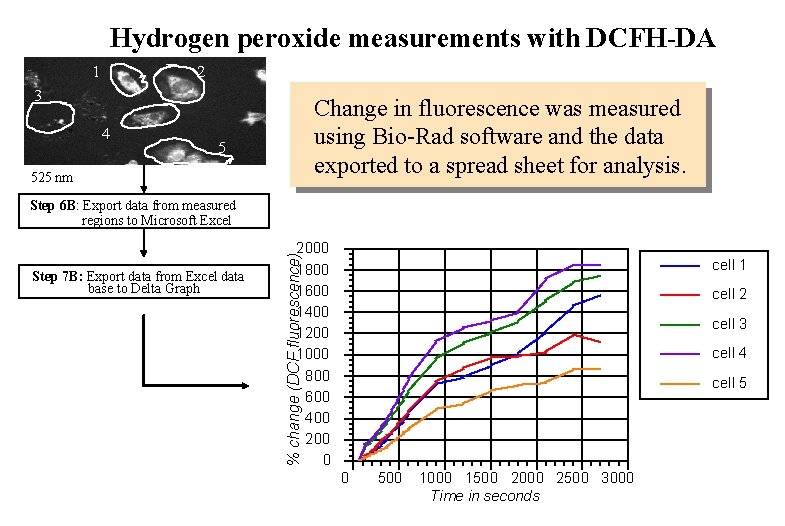
Hydrogen peroxide measurements with DCFH-DA 1 2 3 4 Change in fluorescence was measured using Bio-Rad software and the data exported to a spread sheet for analysis. 5 525 nm Step 6 B: Export data from measured regions to Microsoft Excel % change (DCF fluorescence) Step 7 B: Export data from Excel data base to Delta Graph 2000 1800 1600 1400 1200 1000 800 600 400 200 0 cell 1 cell 2 cell 3 cell 4 cell 5 0 500 1000 1500 2000 2500 3000 Time in seconds
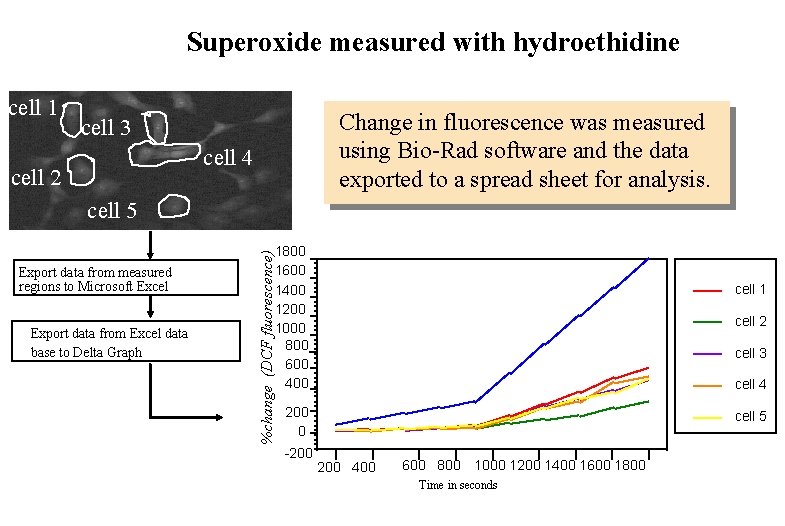
Superoxide measured with hydroethidine cell 1 Change in fluorescence was measured using Bio-Rad software and the data exported to a spread sheet for analysis. cell 3 cell 4 cell 2 cell 5 Export data from Excel data base to Delta Graph %change (DCF fluorescence) Export data from measured regions to Microsoft Excel 1800 1600 1400 1200 1000 800 600 400 cell 1 cell 2 cell 3 cell 4 200 0 -200 cell 5 200 400 600 800 1000 1200 1400 1600 1800 Time in seconds

Promyelocyte Padma Narayanan figures hl 60. ppt Metamyelocyte Myelocyte Neutrophil
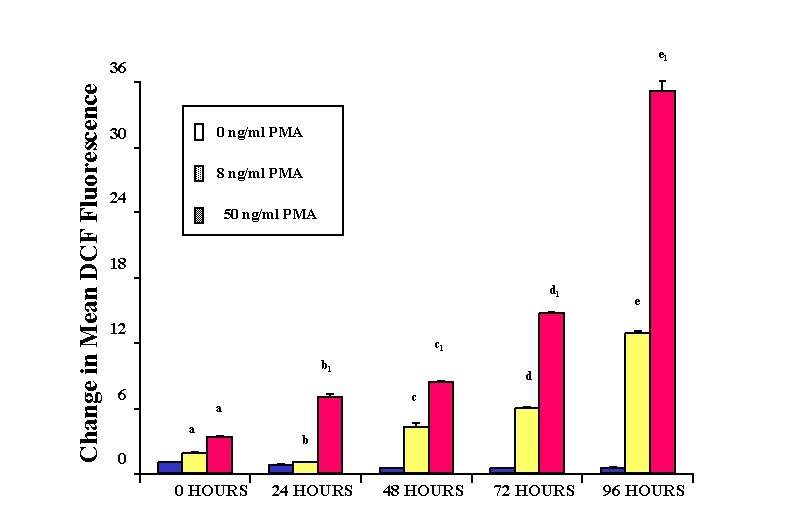
e 1 Change in Mean DCF Fluorescence 36 0 ng/ml PMA 30 8 ng/ml PMA 24 50 ng/ml PMA 18 d 1 12 e c 1 b 1 6 c a a d b 0 0 HOURS 24 HOURS 48 HOURS 72 HOURS 96 HOURS
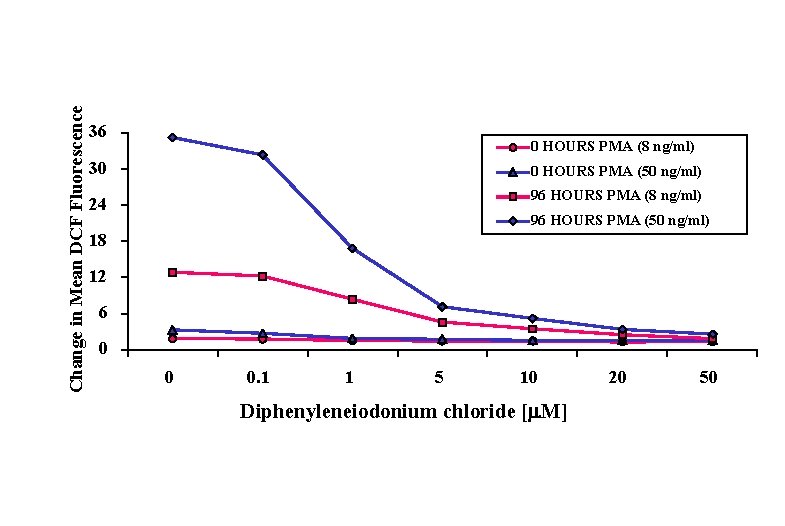
Change in Mean DCF Fluorescence 36 0 HOURS PMA (8 ng/ml) 30 0 HOURS PMA (50 ng/ml) 24 96 HOURS PMA (8 ng/ml) 96 HOURS PMA (50 ng/ml) 18 12 6 0 0 0. 1 1 5 10 Diphenyleneiodonium chloride [m. M] 20 50
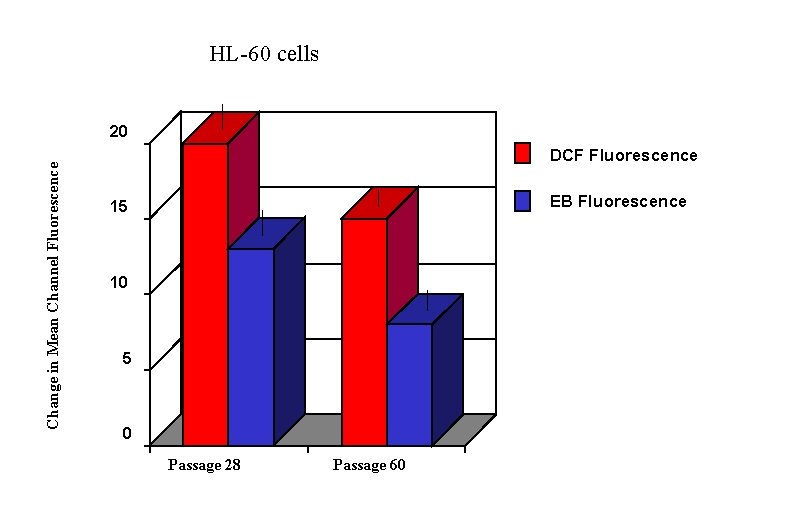
HL-60 cells Change in Mean Channel Fluorescence 20 DCF Fluorescence EB Fluorescence 15 10 5 0 Passage 28 Passage 60
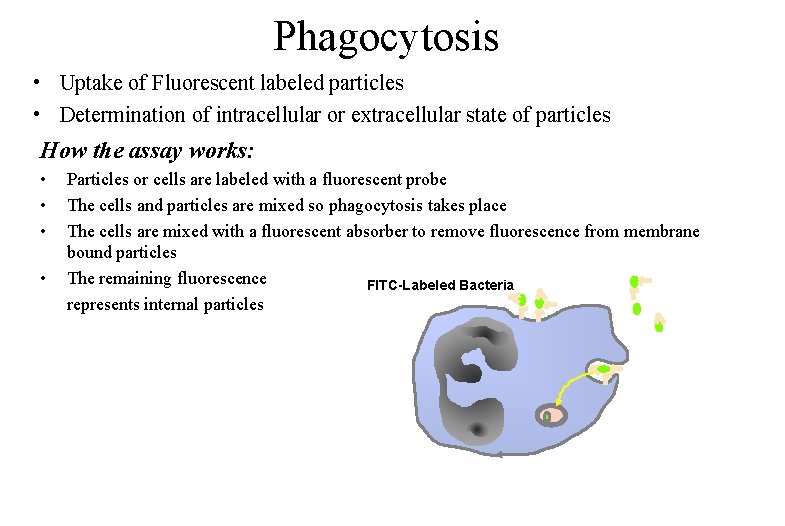
Phagocytosis • Uptake of Fluorescent labeled particles • Determination of intracellular or extracellular state of particles How the assay works: • • Particles or cells are labeled with a fluorescent probe The cells and particles are mixed so phagocytosis takes place The cells are mixed with a fluorescent absorber to remove fluorescence from membrane bound particles The remaining fluorescence FITC-Labeled Bacteria represents internal particles
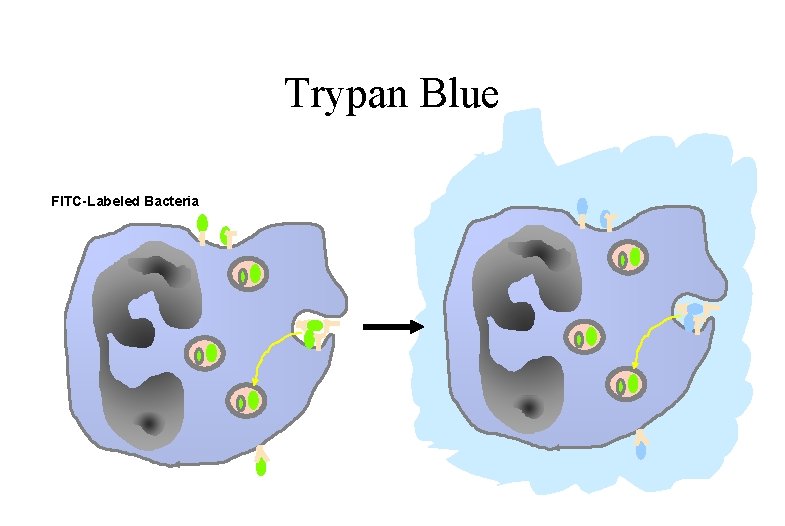
Trypan Blue FITC-Labeled Bacteria
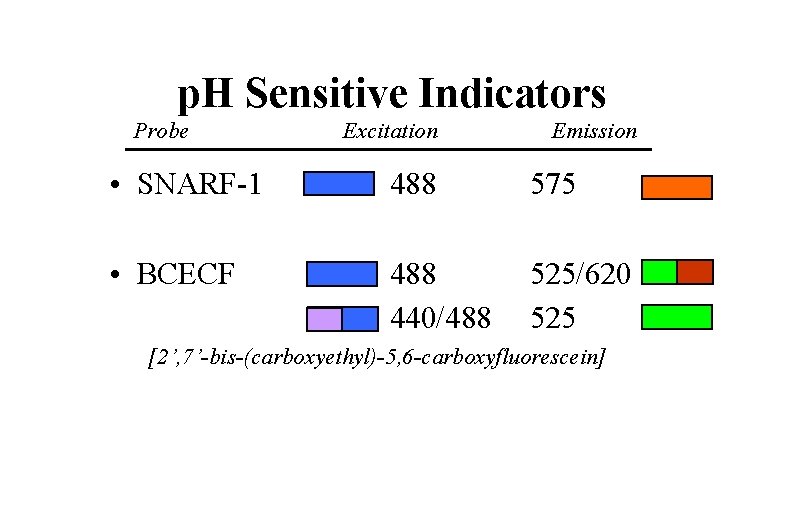
p. H Sensitive Indicators Probe Excitation Emission • SNARF-1 488 575 • BCECF 488 440/488 525/620 525 [2’, 7’-bis-(carboxyethyl)-5, 6 -carboxyfluorescein]
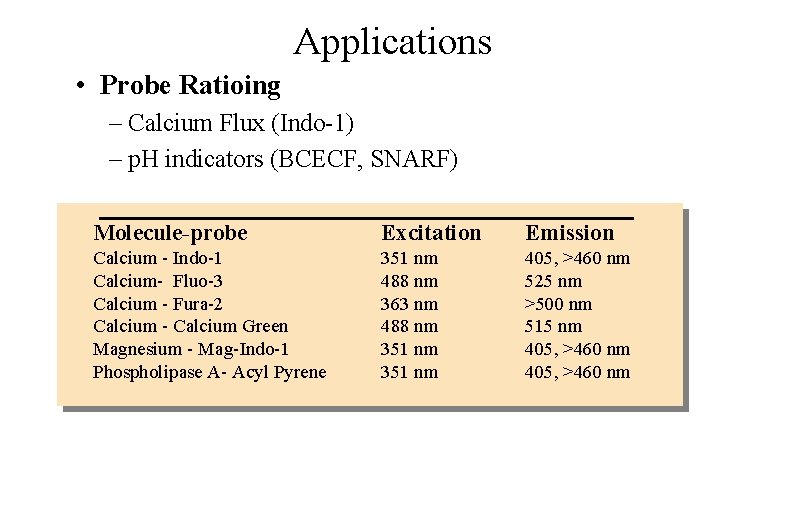
Applications • Probe Ratioing – Calcium Flux (Indo-1) – p. H indicators (BCECF, SNARF) Molecule-probe Excitation Emission Calcium - Indo-1 Calcium- Fluo-3 Calcium - Fura-2 Calcium - Calcium Green Magnesium - Mag-Indo-1 Phospholipase A- Acyl Pyrene 351 nm 488 nm 363 nm 488 nm 351 nm 405, >460 nm 525 nm >500 nm 515 nm 405, >460 nm

Ionic Flux Determinations • Calcium Indo-1 • Intracellular p. H BCECF How the assay works: • Fluorescent probes such as Indo-1 are able to bind to calcium in a ratiometric manner • The emission wavelength decreases as the probe binds available calcium Ratio: intensity of 460 nm / 405 nm signals 0. 8 0. 7 0. 6 0. 5 0. 4 0. 3 0. 2 0. 1 Stimulation 0 36 72 108 Time (Seconds) 144 180 Flow Cytometry 0 0 Image Analysis 50 Time (seconds) 100 150 200
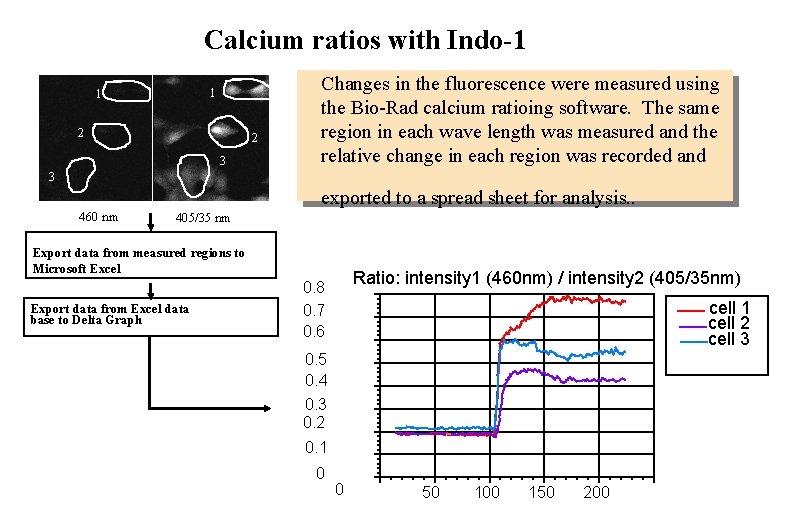
Calcium ratios with Indo-1 1 1 2 2 3 Changes in the fluorescence were measured using the Bio-Rad calcium ratioing software. The same region in each wave length was measured and the relative change in each region was recorded and 3 exported to a spread sheet for analysis. . 460 nm 405/35 nm Export data from measured regions to Microsoft Excel Export data from Excel data base to Delta Graph Ratio: intensity 1 (460 nm) / intensity 2 (405/35 nm) 0. 8 0. 7 0. 6 cell 1 cell 2 cell 3 0. 5 0. 4 0. 3 0. 2 0. 1 0 0 50 100 150 200
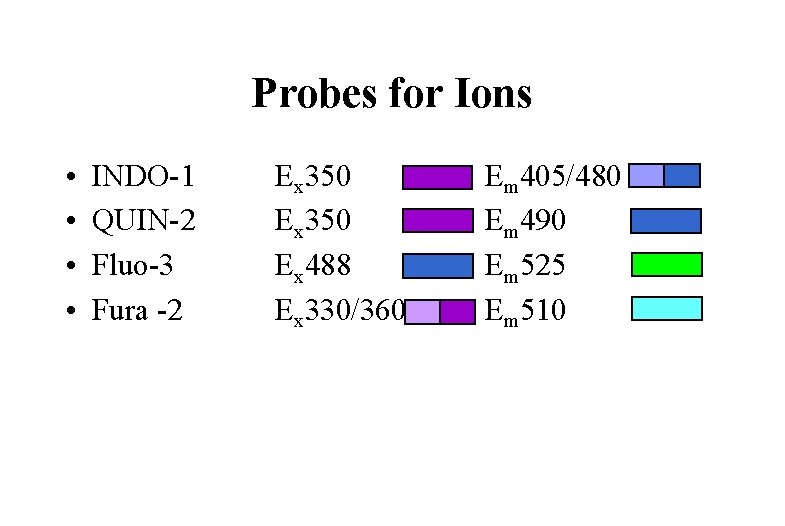
Probes for Ions • • INDO-1 QUIN-2 Fluo-3 Fura -2 Ex 350 Ex 488 Ex 330/360 Em 405/480 Em 490 Em 525 Em 510
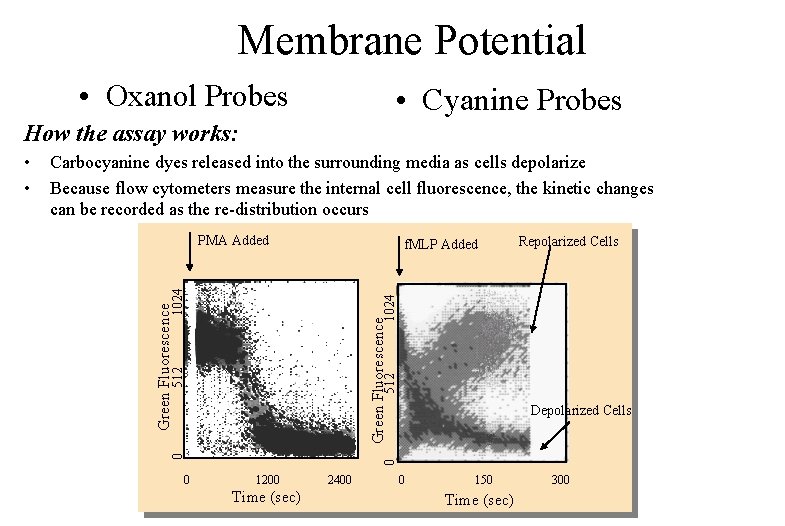
Membrane Potential • Oxanol Probes • Cyanine Probes How the assay works: Carbocyanine dyes released into the surrounding media as cells depolarize Because flow cytometers measure the internal cell fluorescence, the kinetic changes can be recorded as the re-distribution occurs PMA Added 512 Green Fluorescence 0 Repolarized Cells 1024 f. MLP Added Depolarized Cells 0 0 • • 1200 Time (sec) 2400 0 150 Time (sec) 300

Lipid Peroxidation • Probe: 5 M cis-paranaric acid (Molecular Probes) How the assay works: • Cis-paranaric acid is a naturally fluorescent fatty acid which has 4 conjugated double bonds which become targeted by lipid peroxidation reactions with a subsequent loss of fluorescence
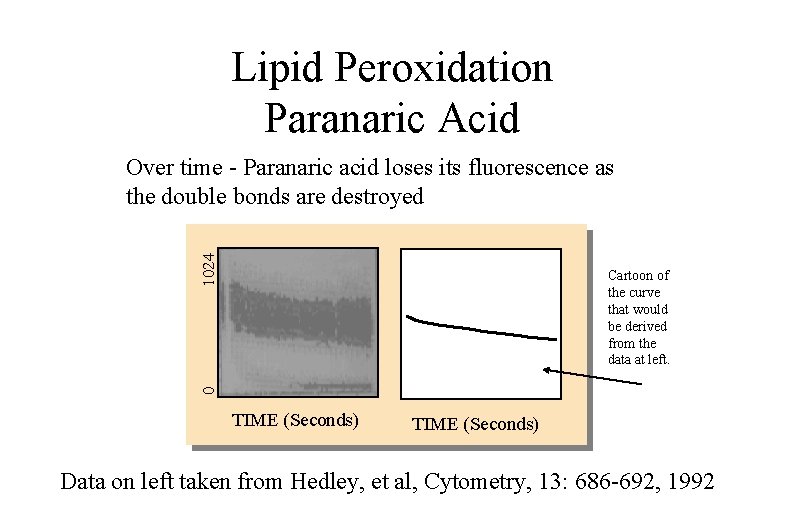
Lipid Peroxidation Paranaric Acid 1024 Over time - Paranaric acid loses its fluorescence as the double bonds are destroyed 0 Cartoon of the curve that would be derived from the data at left. TIME (Seconds) Data on left taken from Hedley, et al, Cytometry, 13: 686 -692, 1992
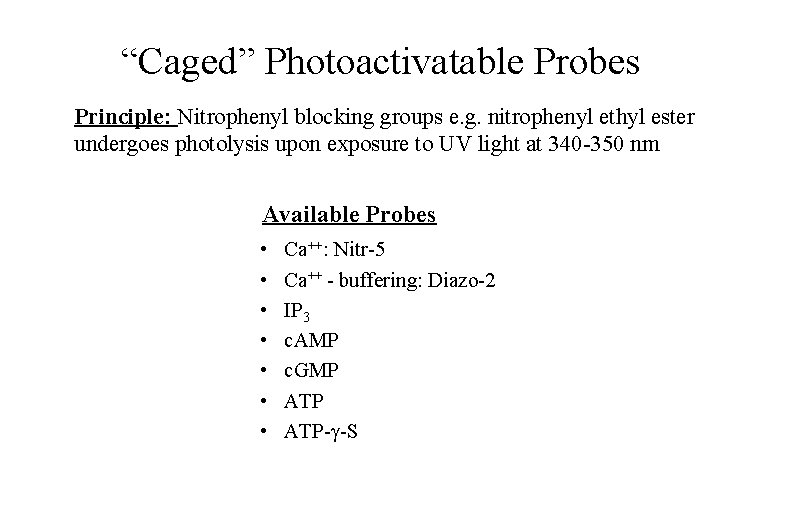
“Caged” Photoactivatable Probes Principle: Nitrophenyl blocking groups e. g. nitrophenyl ethyl ester undergoes photolysis upon exposure to UV light at 340 -350 nm Available Probes • • Ca++: Nitr-5 Ca++ - buffering: Diazo-2 IP 3 c. AMP c. GMP ATP- -S
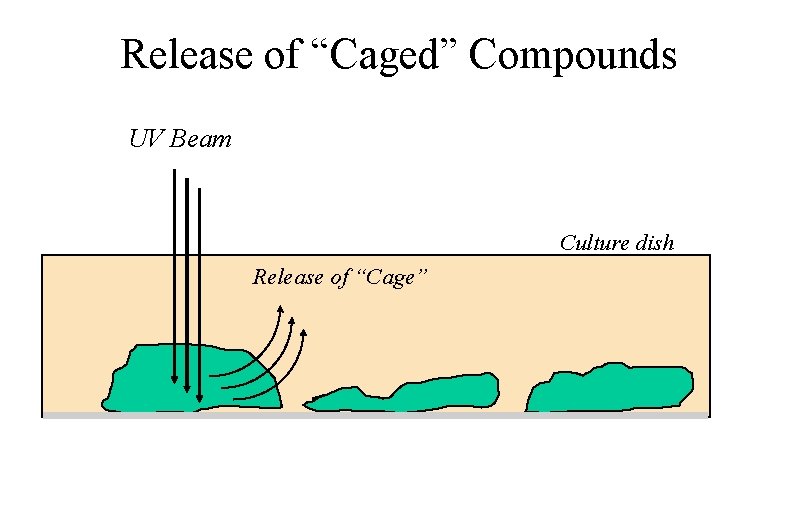
Release of “Caged” Compounds UV Beam Culture dish Release of “Cage”
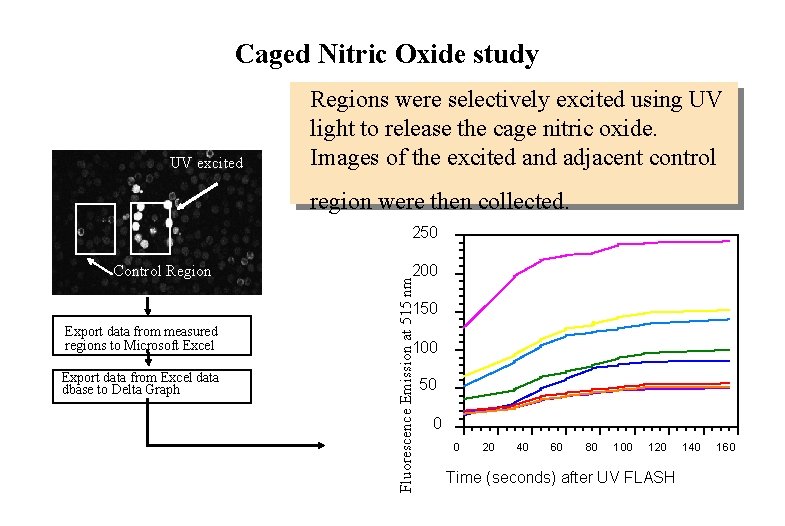
Caged Nitric Oxide study UV excited Regions were selectively excited using UV light to release the cage nitric oxide. Images of the excited and adjacent control region were then collected. 250 200 Fluorescence Emission at 515 nm Control Region 150 Export data from measured regions to Microsoft Excel Export data from Excel data dbase to Delta Graph 100 50 0 0 20 40 60 80 100 0 120 Time (seconds) after UV FLASH 140 160
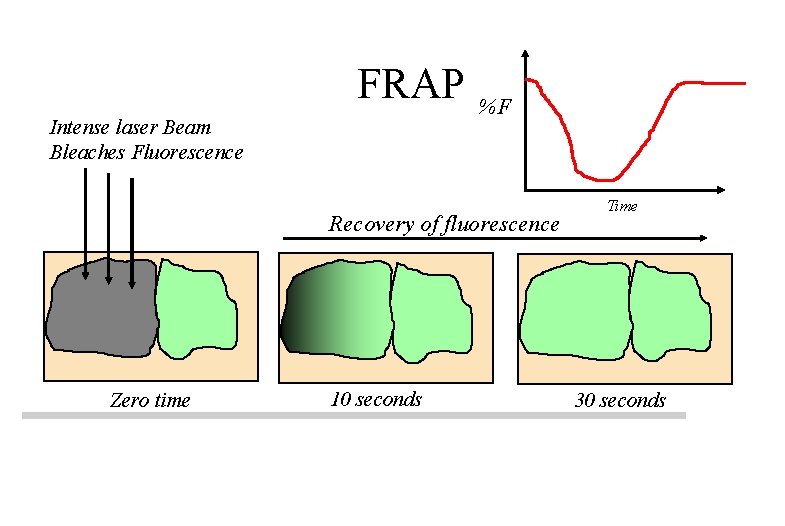
FRAP Intense laser Beam Bleaches Fluorescence %F Recovery of fluorescence Zero time 10 seconds Time 30 seconds
Conclusions & Summary Functional Studies In Cytometry • • • Oxygen radicals Nitrogen radicals Antioxidants Cell viability Organelle function • • Lipid peroxidation Membrane potential Calcium fluxes p. H changes

Acknowledgements • • • Kathy Ragheb Gretchen Lawler Steve Kelley Monica Shively Dave Whittinghill Stephanie Sincock Karen Cornell Karin Kooreman Nian-Yu Li Padma Narayanan (Smith Kline) Wayne Carter (Pfizer)
- Slides: 43