PROTEOMICS Mass spectrometry in Biochemistry Unit 3 LCMS
PROTEOMICS (Mass spectrometry in Biochemistry) Unit 3: LC-MS, quantitation, biomarkers, ion mobility MS, 3 D structure (HDX)
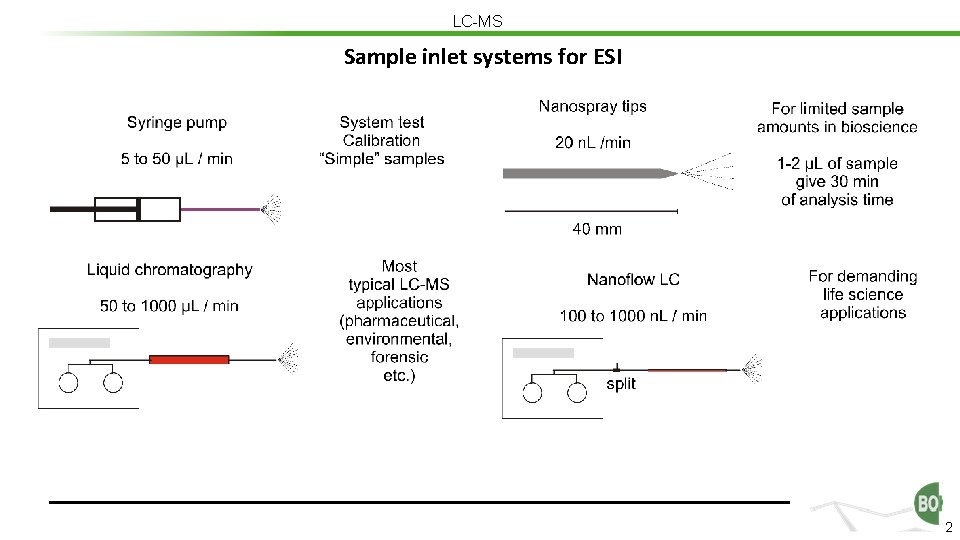
LC-MS Sample inlet systems for ESI 2
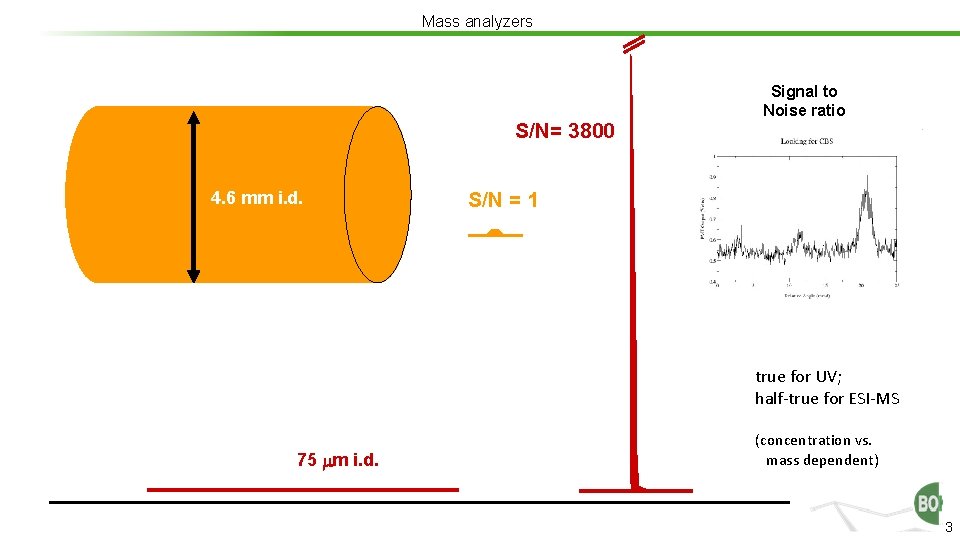
Mass analyzers S/N= 3800 4. 6 mm i. d. Signal to Noise ratio S/N = 1 1. 0 ml/min true for UV; half-true for ESI-MS 75 m i. d. (concentration vs. mass dependent) 3
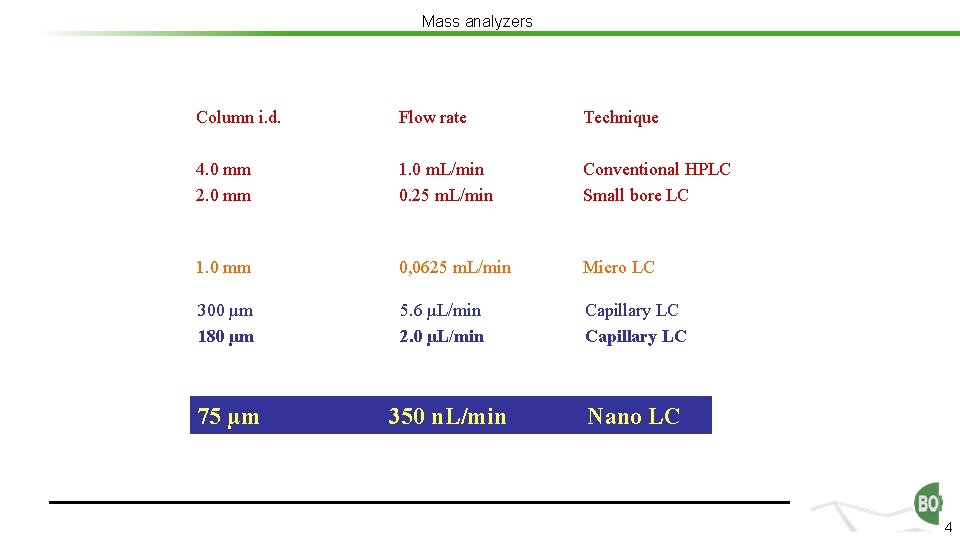
Mass analyzers Column i. d. Flow rate Technique 4. 0 mm 2. 0 mm 1. 0 m. L/min 0. 25 m. L/min Conventional HPLC Small bore LC 1. 0 mm 0, 0625 m. L/min Micro LC 300 µm 180 µm 5. 6 µL/min 2. 0 µL/min Capillary LC 75 µm 350 n. L/min Nano LC 4
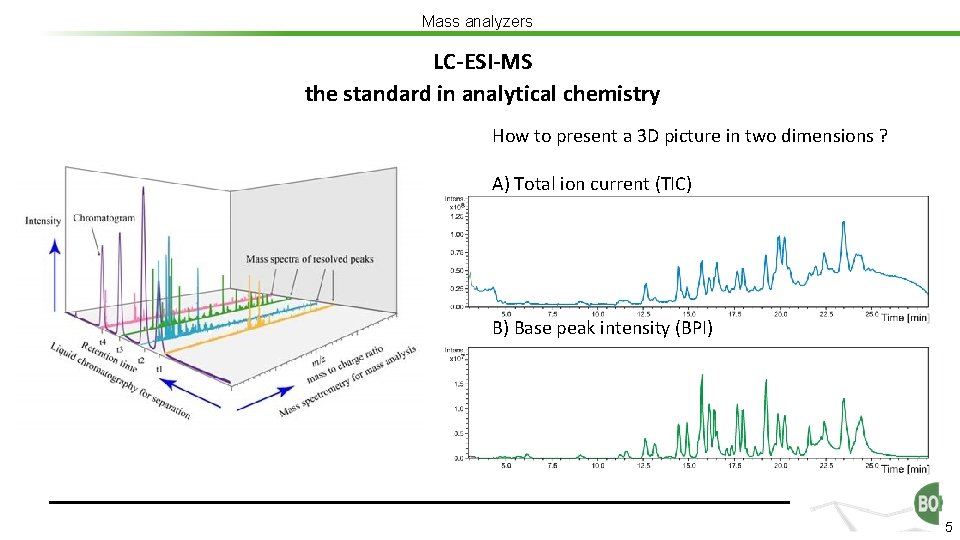
Mass analyzers LC-ESI-MS the standard in analytical chemistry How to present a 3 D picture in two dimensions ? A) Total ion current (TIC) B) Base peak intensity (BPI) 5
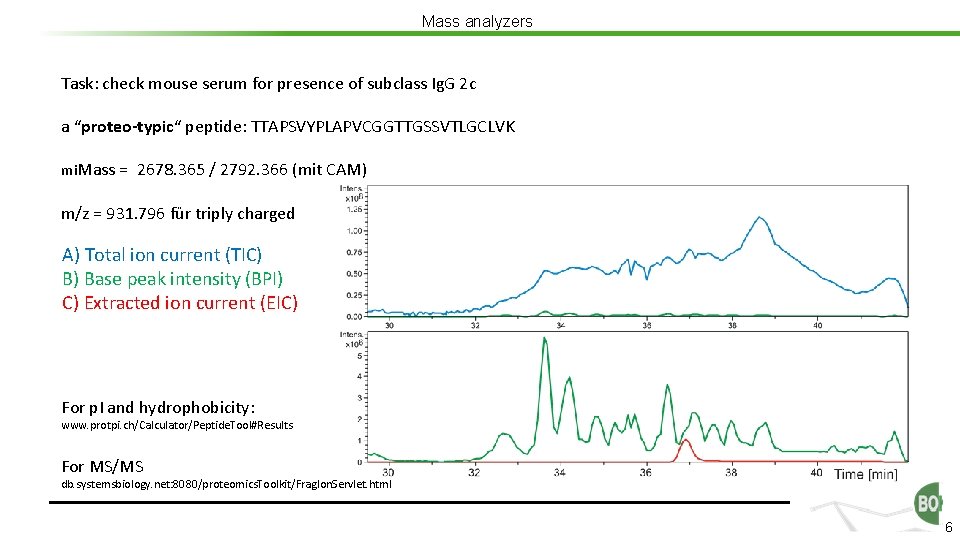
Mass analyzers Task: check mouse serum for presence of subclass Ig. G 2 c a “proteo-typic“ peptide: TTAPSVYPLAPVCGGTTGSSVTLGCLVK mi. Mass = 2678. 365 / 2792. 366 (mit CAM) m/z = 931. 796 für triply charged A) Total ion current (TIC) B) Base peak intensity (BPI) C) Extracted ion current (EIC) For p. I and hydrophobicity: www. protpi. ch/Calculator/Peptide. Tool#Results For MS/MS db. systemsbiology. net: 8080/proteomics. Toolkit/Frag. Ion. Servlet. html 6
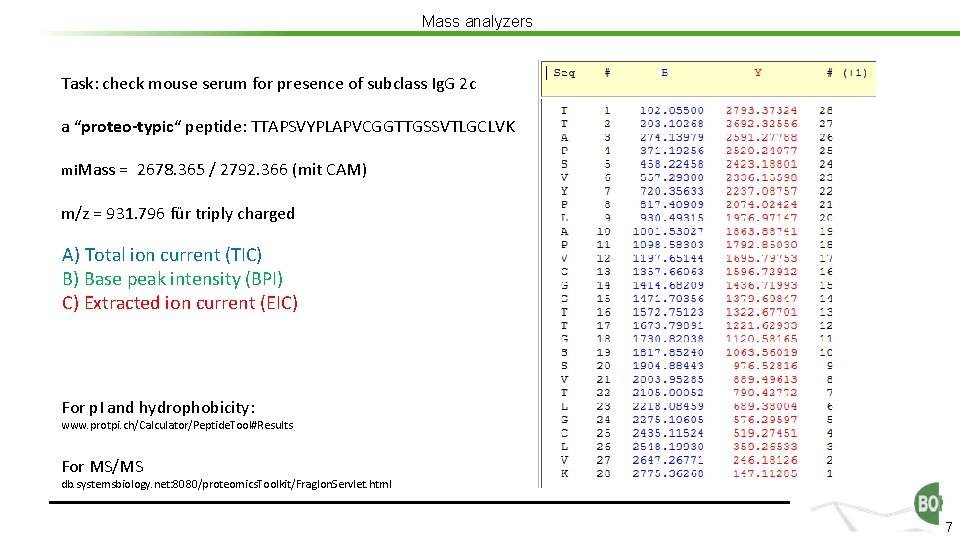
Mass analyzers Task: check mouse serum for presence of subclass Ig. G 2 c a “proteo-typic“ peptide: TTAPSVYPLAPVCGGTTGSSVTLGCLVK mi. Mass = 2678. 365 / 2792. 366 (mit CAM) m/z = 931. 796 für triply charged A) Total ion current (TIC) B) Base peak intensity (BPI) C) Extracted ion current (EIC) For p. I and hydrophobicity: www. protpi. ch/Calculator/Peptide. Tool#Results For MS/MS db. systemsbiology. net: 8080/proteomics. Toolkit/Frag. Ion. Servlet. html 7
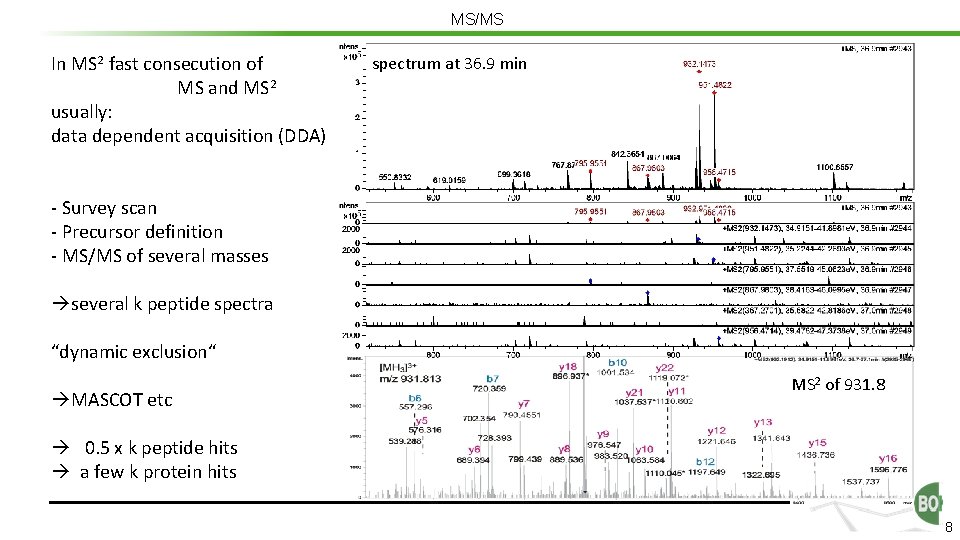
MS/MS In MS 2 fast consecution of MS and MS 2 usually: data dependent acquisition (DDA) spectrum at 36. 9 min - Survey scan - Precursor definition - MS/MS of several masses several k peptide spectra “dynamic exclusion“ MASCOT etc MS 2 of 931. 8 0. 5 x k peptide hits a few k protein hits 8
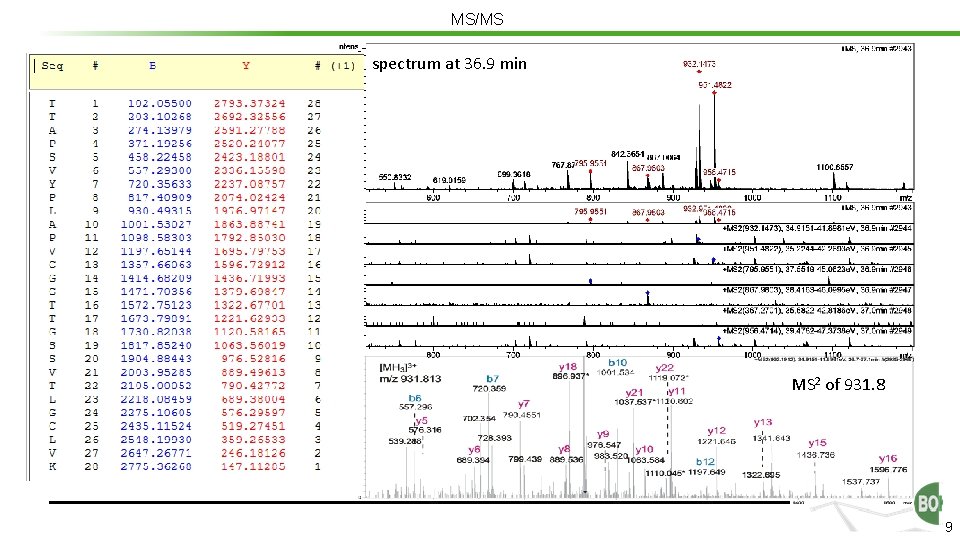
MS/MS spectrum at 36. 9 min MS 2 of 931. 8 9
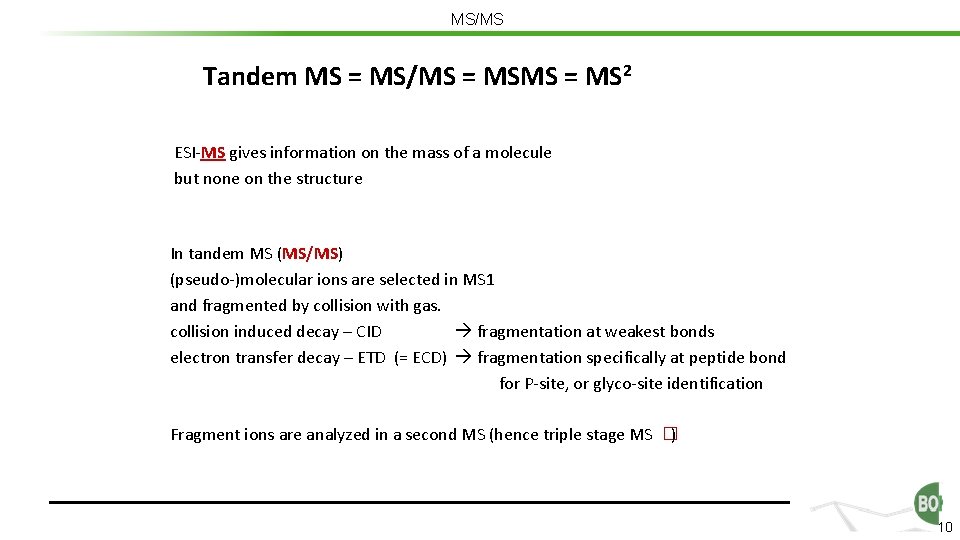
MS/MS Tandem MS = MS/MS = MS 2 ESI-MS gives information on the mass of a molecule but none on the structure In tandem MS (MS/MS) (pseudo-)molecular ions are selected in MS 1 and fragmented by collision with gas. collision induced decay – CID fragmentation at weakest bonds electron transfer decay – ETD (= ECD) fragmentation specifically at peptide bond for P-site, or glyco-site identification Fragment ions are analyzed in a second MS (hence triple stage MS �) 10
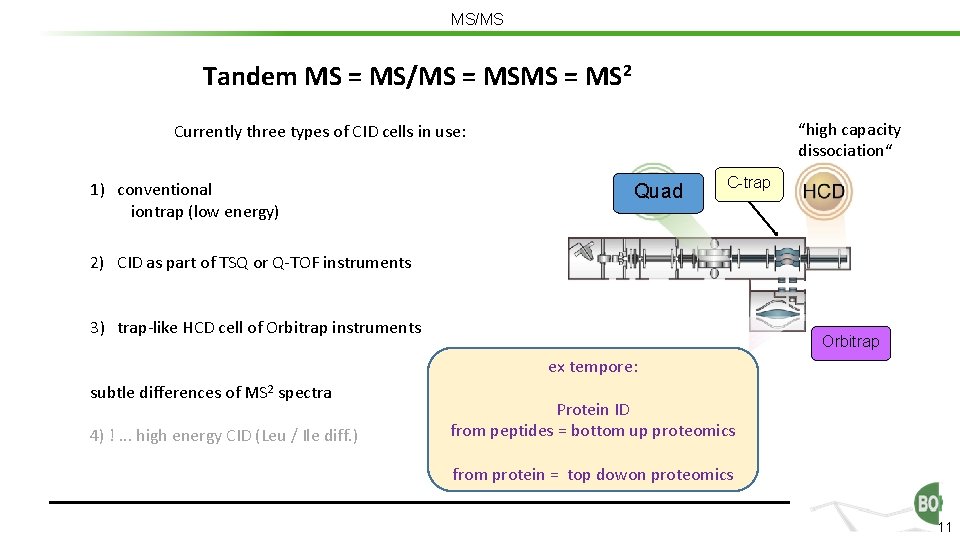
MS/MS Tandem MS = MS/MS = MS 2 “high capacity dissociation“ Currently three types of CID cells in use: 1) conventional iontrap (low energy) Quad C-trap 2) CID as part of TSQ or Q-TOF instruments 3) trap-like HCD cell of Orbitrap instruments Orbitrap ex tempore: subtle differences of MS 2 spectra 4) !. . . high energy CID (Leu / Ile diff. ) Protein ID from peptides = bottom up proteomics from protein = top dowon proteomics 11
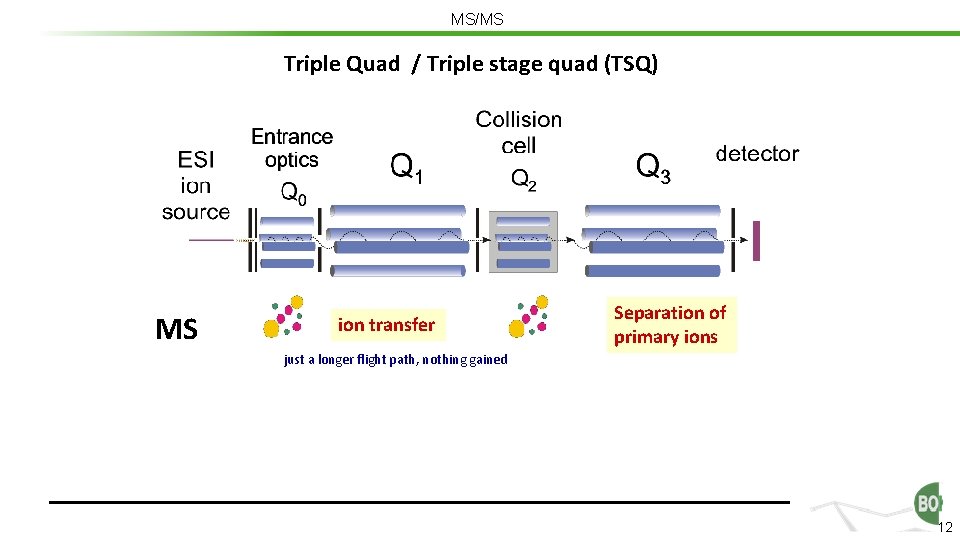
MS/MS Triple Quad / Triple stage quad (TSQ) MS ion transfer Separation of primary ions just a longer flight path, nothing gained 12
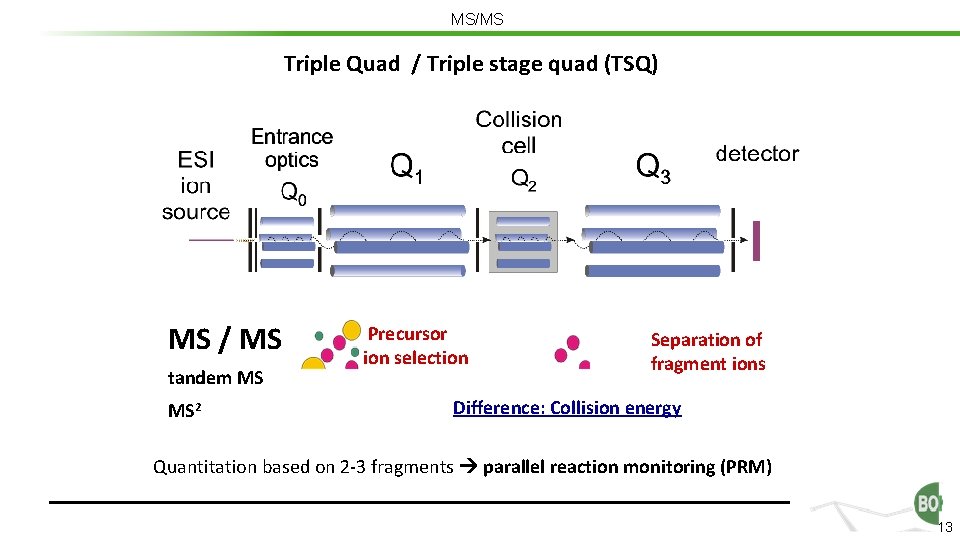
MS/MS Triple Quad / Triple stage quad (TSQ) MS / MS tandem MS MS 2 Precursor ion selection Separation of fragment ions Difference: Collision energy Quantitation based on 2 -3 fragments parallel reaction monitoring (PRM) 13
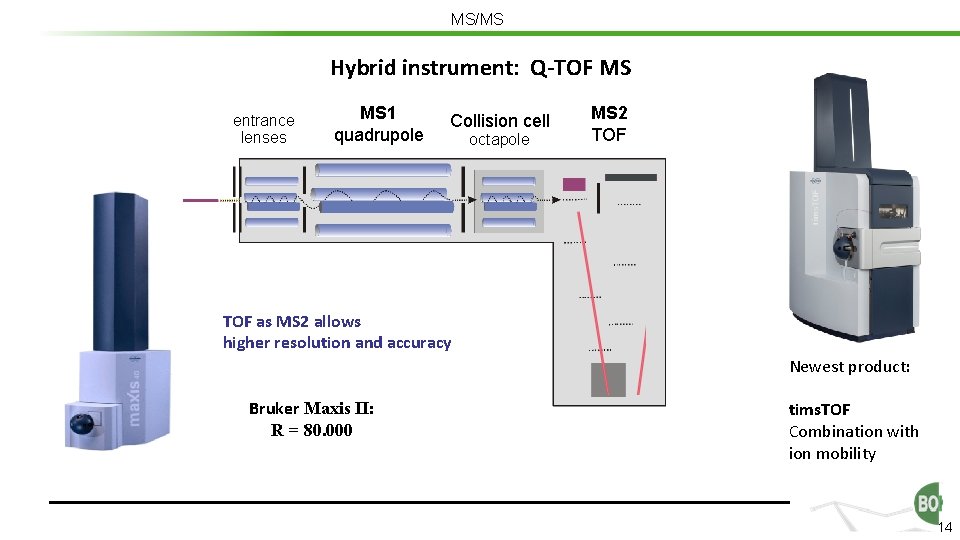
MS/MS Hybrid instrument: Q-TOF MS entrance lenses MS 1 quadrupole Collision cell octapole MS 2 TOF as MS 2 allows higher resolution and accuracy Newest product: Bruker Maxis II: R = 80. 000 tims. TOF Combination with ion mobility 14
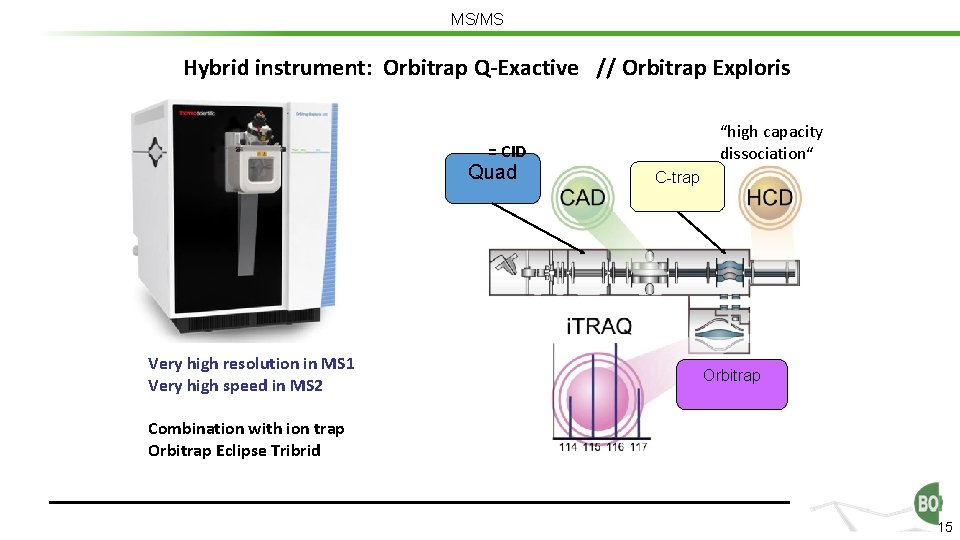
MS/MS Hybrid instrument: Orbitrap Q-Exactive // Orbitrap Exploris “high capacity dissociation“ = CID Quad Very high resolution in MS 1 Very high speed in MS 2 C-trap Orbitrap Combination with ion trap Orbitrap Eclipse Tribrid 15
MS/MS in proteomics Triple-quad resolution insufficient for exact precursor mass determination Q-TOF Q-Orbitrap (Q-Exactive, Orbitrap Exploris) IT-Orbitrap (Orbitrap Fusion Tribrid etc. ) how to increase id quality and proteome coverage ? strategy I more peptides (see next page) strategy II knowledge based (SWATH etc. ) relevant: good chromatography UPLC on long columns, > 5000 proteins in one run 16
MS/MS in proteomics Triple-quad resolution insufficient for exact precursor mass determination Q-TOF Q-Orbitrap (Q-Exactive, Orbitrap Exploris) IT-Orbitrap (Orbitrap Fusion Tribrid etc. ) how to increase id quality and proteome coverage ? strategy I more peptides (see next page) strategy II knowledge based (SWATH etc. ) 17
MS/MS in proteomics how to increase id quality and proteome coverage ? strategy I more peptides history: two-dimensional LC (aka Mud. PIT, multi-dimensional protein identification technology) 1. dim: cation X - 2. dim: RP at acidic p. H � 2 1. dim: RP at ph � 9 - 2. dim: RP at acidic p. H � 2 topical: good chromatography = UPLC on long columns 5000 proteins in one run fast and sensitive instruments 18
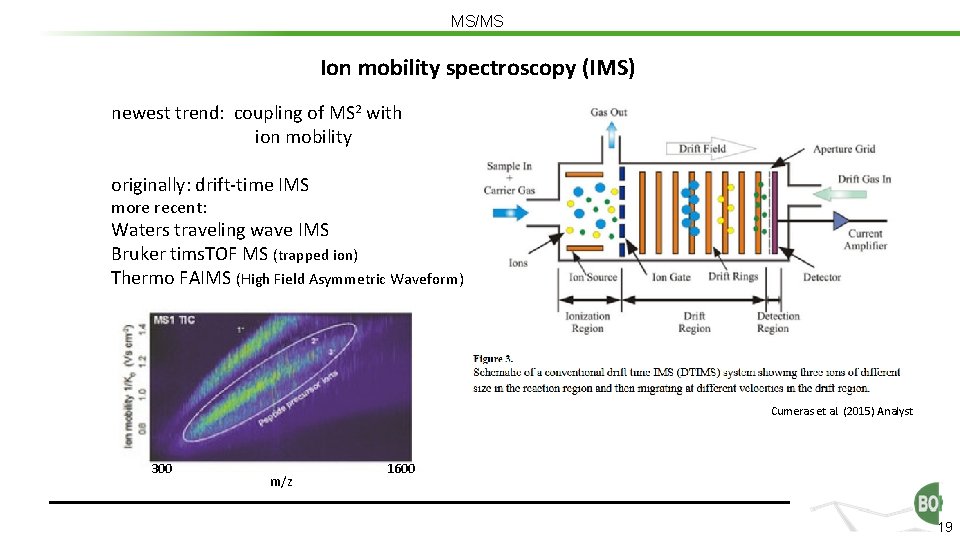
MS/MS Ion mobility spectroscopy (IMS) newest trend: coupling of MS 2 with ion mobility originally: drift-time IMS more recent: Waters traveling wave IMS Bruker tims. TOF MS (trapped ion) Thermo FAIMS (High Field Asymmetric Waveform) Cumeras et al. (2015) Analyst 300 m/z 1600 19
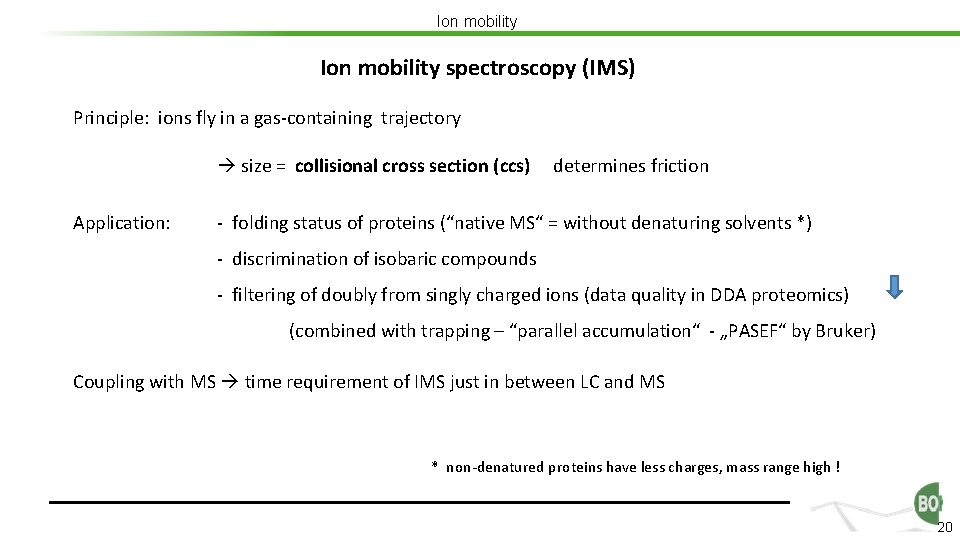
Ion mobility spectroscopy (IMS) Principle: ions fly in a gas-containing trajectory size = collisional cross section (ccs) Application: determines friction - folding status of proteins (“native MS“ = without denaturing solvents *) - discrimination of isobaric compounds - filtering of doubly from singly charged ions (data quality in DDA proteomics) (combined with trapping – “parallel accumulation“ - „PASEF“ by Bruker) Coupling with MS time requirement of IMS just in between LC and MS * non-denatured proteins have less charges, mass range high ! 20
Quantitation in MS Quantitation !! Peak height does not reliably reflect amount / concentration (different ionization and transfer efficiency, matrix effects etc. Comparison of apples and apples is possible ! Options: Label-free quantitation inject sample A inject sample B Use of stable isotopes label 2 – 8 samples with different masses, mix, analyze in one run What is the aim: Absolute or relative quantitation ? 21
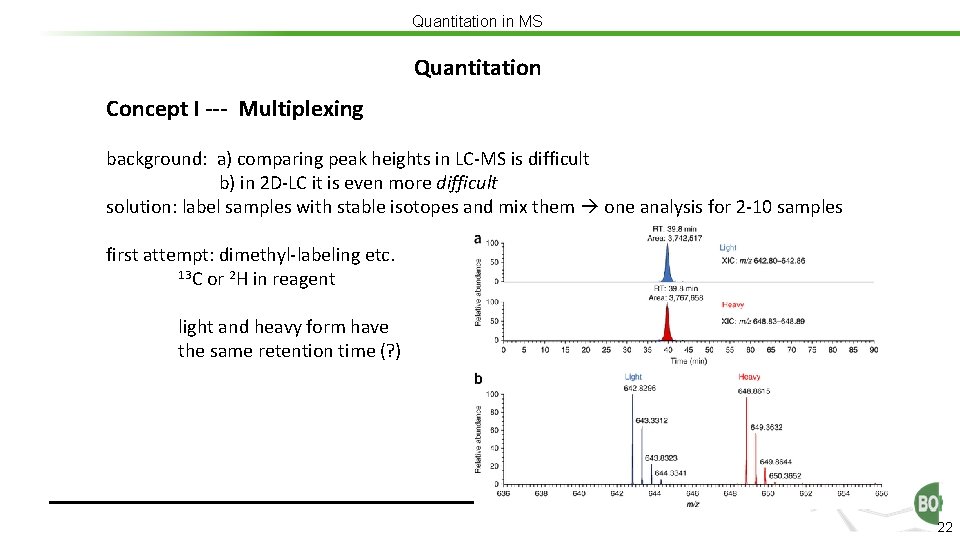
Quantitation in MS Quantitation Concept I --- Multiplexing background: a) comparing peak heights in LC-MS is difficult b) in 2 D-LC it is even more difficult solution: label samples with stable isotopes and mix them one analysis for 2 -10 samples first attempt: dimethyl-labeling etc. 13 C or 2 H in reagent light and heavy form have the same retention time (? ) 22
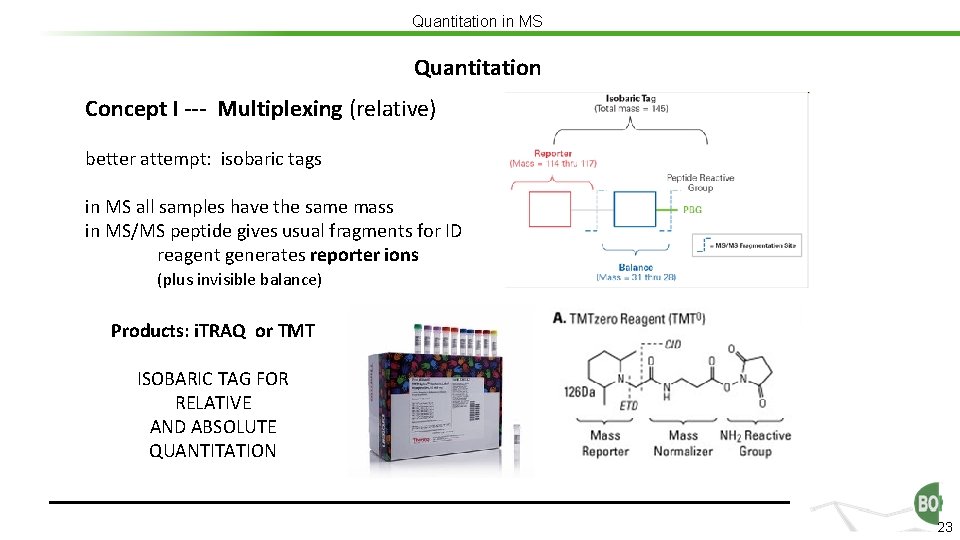
Quantitation in MS Quantitation Concept I --- Multiplexing (relative) better attempt: isobaric tags in MS all samples have the same mass in MS/MS peptide gives usual fragments for ID reagent generates reporter ions (plus invisible balance) Products: i. TRAQ or TMT ISOBARIC TAG FOR RELATIVE AND ABSOLUTE QUANTITATION 23
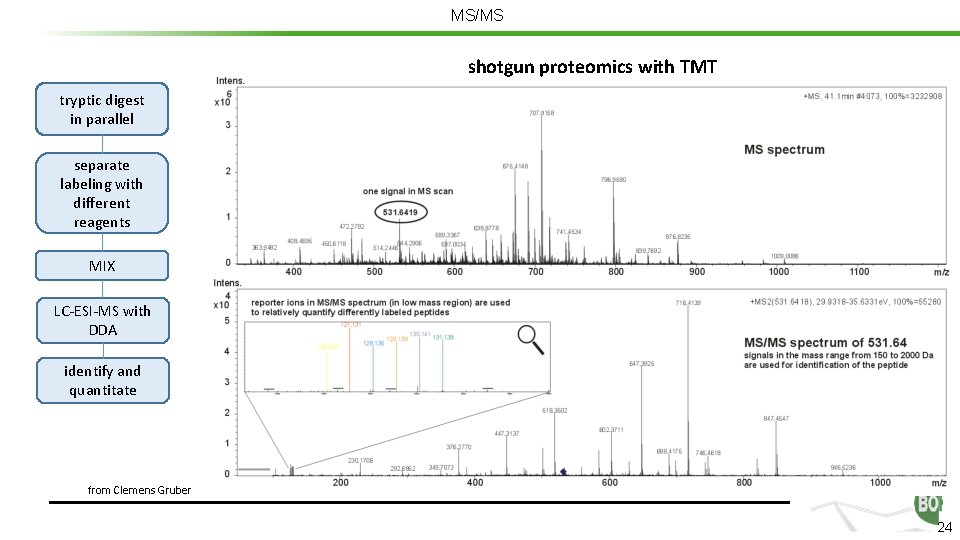
MS/MS shotgun proteomics with TMT tryptic digest in parallel separate labeling with different reagents MIX LC-ESI-MS with DDA identify and quantitate from Clemens Gruber 24
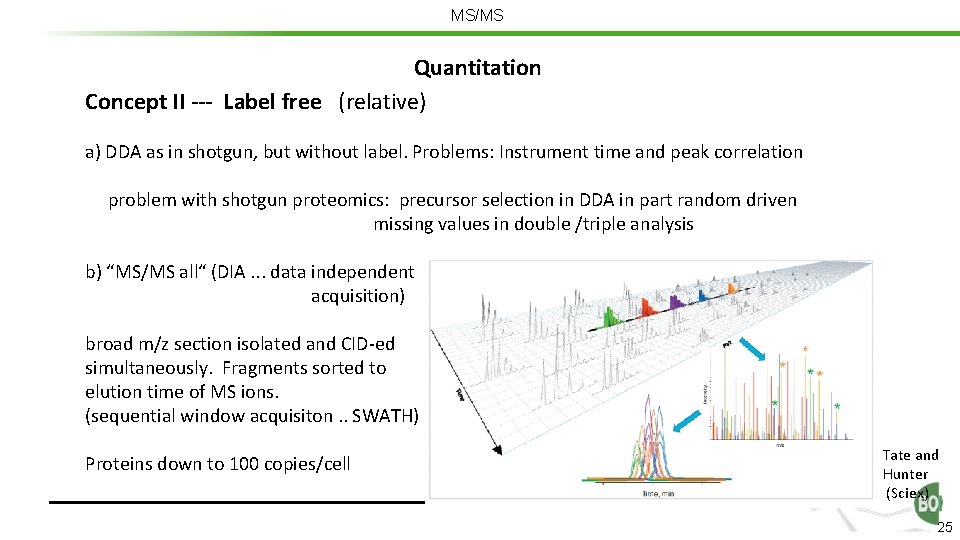
MS/MS Quantitation Concept II --- Label free (relative) a) DDA as in shotgun, but without label. Problems: Instrument time and peak correlation problem with shotgun proteomics: precursor selection in DDA in part random driven missing values in double /triple analysis b) “MS/MS all“ (DIA. . . data independent acquisition) broad m/z section isolated and CID-ed simultaneously. Fragments sorted to elution time of MS ions. (sequential window acquisiton. . SWATH) Proteins down to 100 copies/cell Tate and Hunter (Sciex) 25
MS/MS Quantitation Concept III --- with „internal standard“ (absolute) proteotypic peptides with heavy isotopes are mixed to sample. . . ACQUA peptides quantitation performed either with TSQ instruments on fragments (low resolution; fixed MS 1 windows, defined MRM masses ) or HR instruments on precursor (exact mass quantitation) in general: analyte is known targeted MS analysis increasing use in clinical diagnosis (substitutes ELISAs) 26
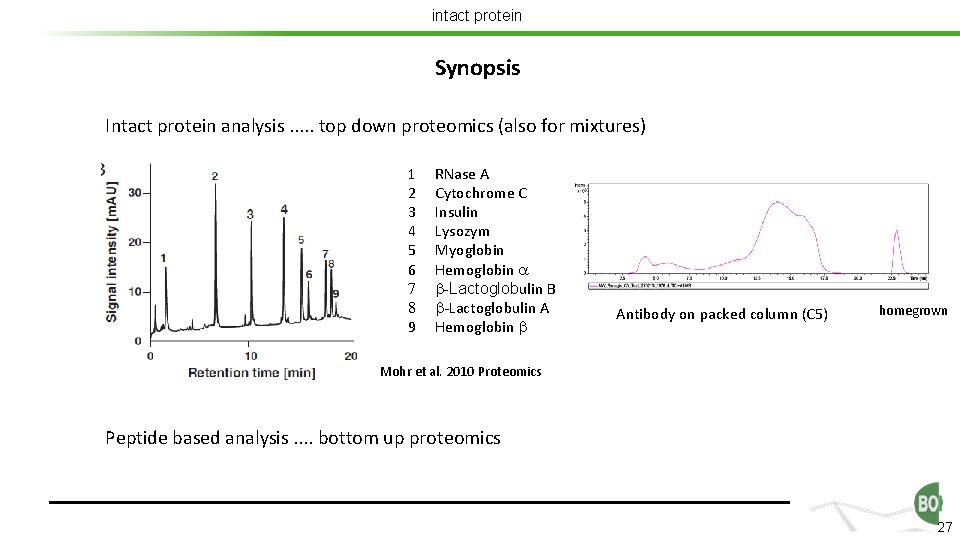
intact protein Synopsis Intact protein analysis. . . top down proteomics (also for mixtures) 1 2 3 4 5 6 7 8 9 RNase A Cytochrome C Insulin Lysozym Myoglobin Hemoglobin a b-Lactoglobulin B b-Lactoglobulin A Hemoglobin b Antibody on packed column (C 5) homegrown Mohr et al. 2010 Proteomics Peptide based analysis. . bottom up proteomics 27
intact protein Synopsis Intact protein analysis. . . top down proteomics (also for mixtures) middle down proteomics www. genovis. com options: reduction of S-S bonds – H- and L-chains cleavage below hinge disulfides – Fab 2, Fc cleavage above hinge disulfides – 2 Fab, fc Peptide based analysis. . bottom up proteomics 28
Tests with a binary result Temperature Weight Height pass / fail healthy / sick Concentration Ratio Income accept / reject …. In medicine: search for biomarkers
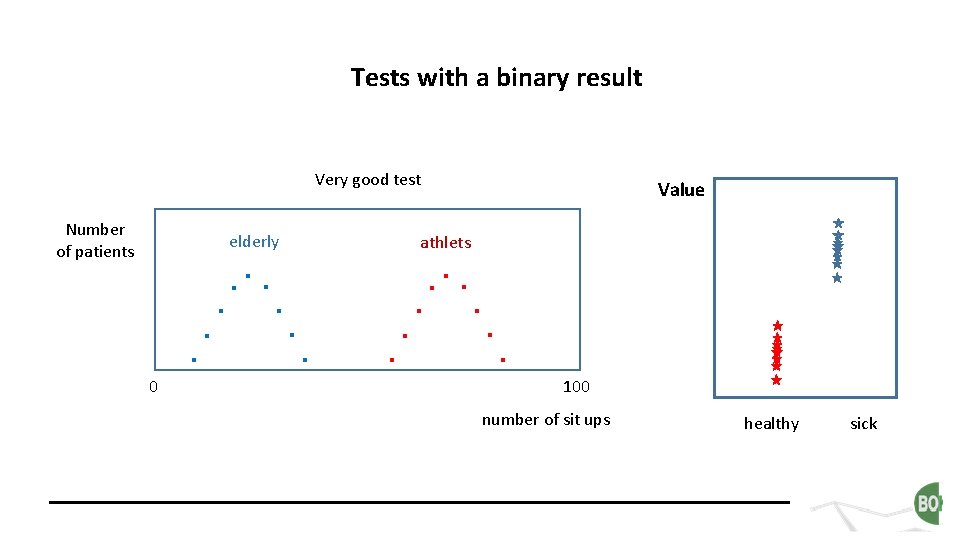
Tests with a binary result Very good test Number of patients elderly . 0 . . . Value athlets . . . 100 number of sit ups healthy sick
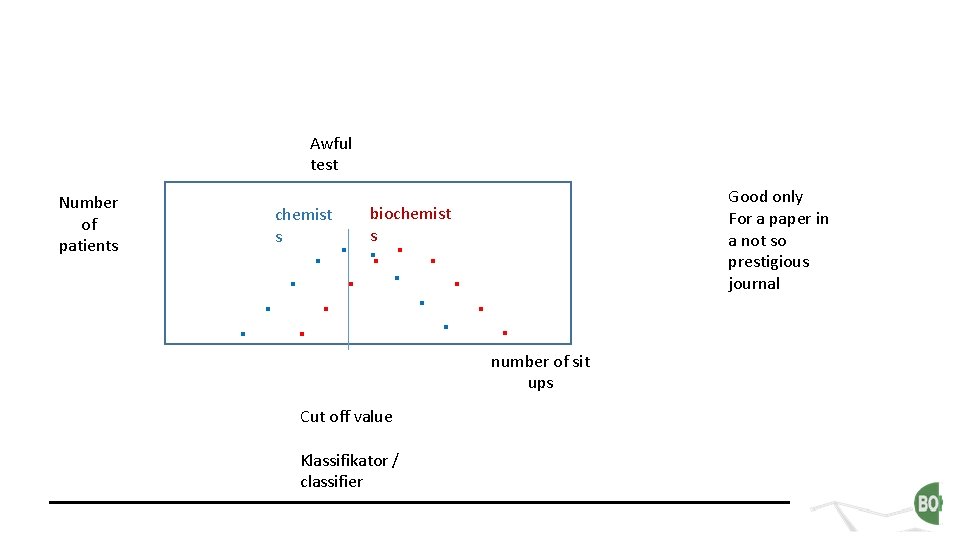
Awful test Number of patients chemist s biochemist s . . . . number of sit ups Cut off value Klassifikator / classifier Good only For a paper in a not so prestigious journal
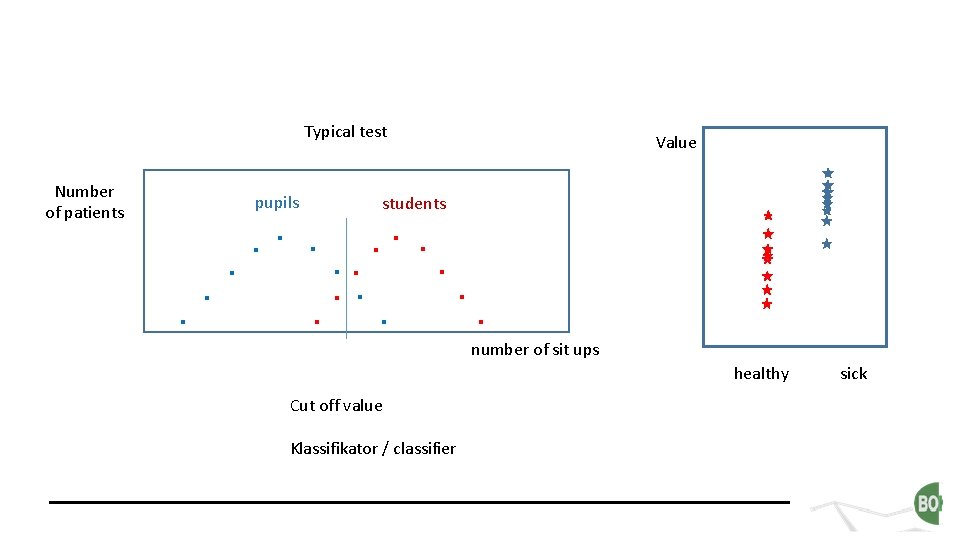
Typical test Number of patients pupils . . . . Value students . . . number of sit ups healthy Cut off value Klassifikator / classifier sick
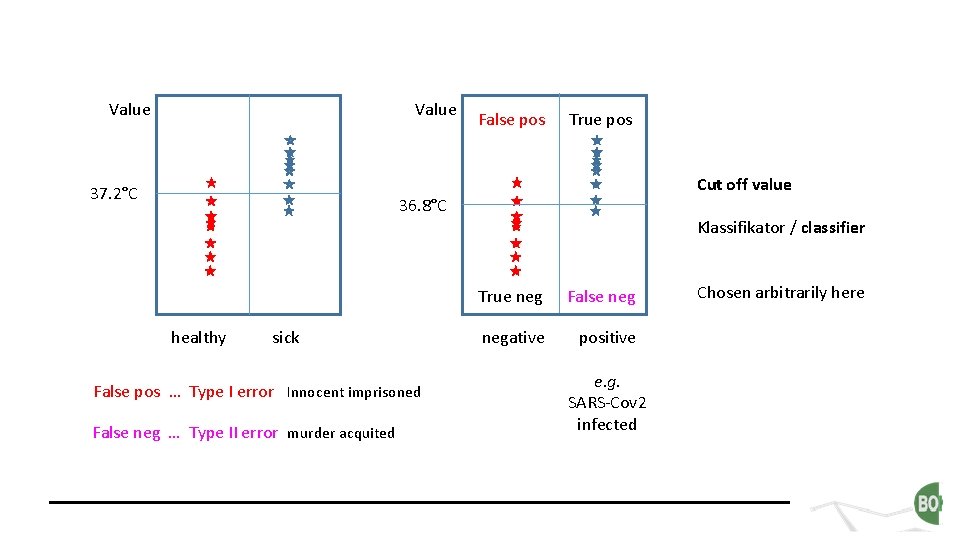
Value 37. 2°C False pos True pos Cut off value 36. 8°C Klassifikator / classifier healthy sick False pos … Type I error Innocent imprisoned False neg … Type II error murder acquited True neg False negative positive e. g. SARS-Cov 2 infected Chosen arbitrarily here
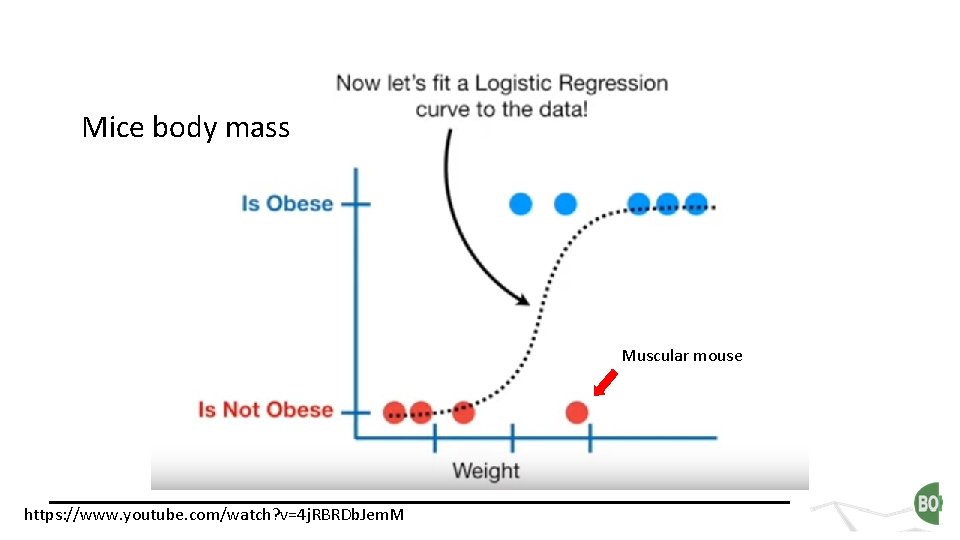
Mice body mass Muscular mouse https: //www. youtube. com/watch? v=4 j. RBRDb. Jem. M
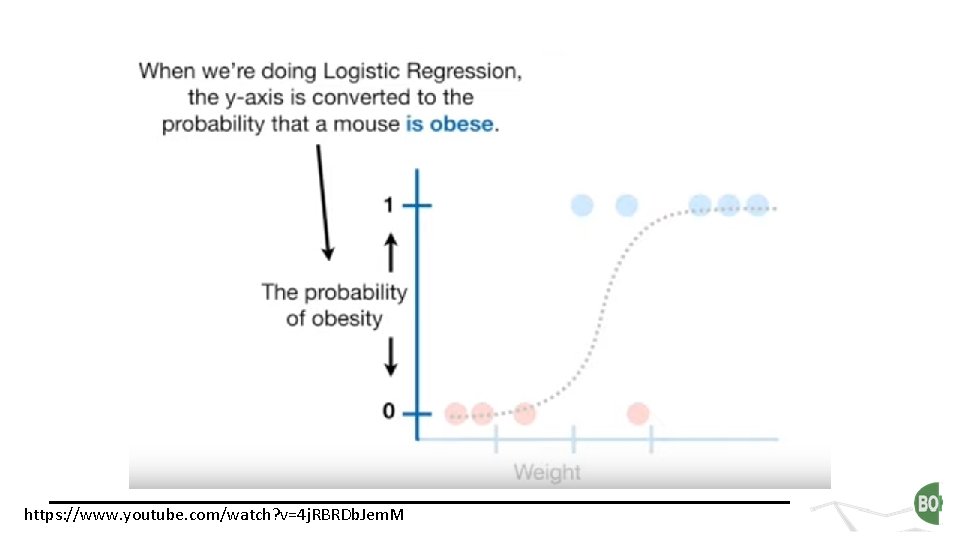
https: //www. youtube. com/watch? v=4 j. RBRDb. Jem. M
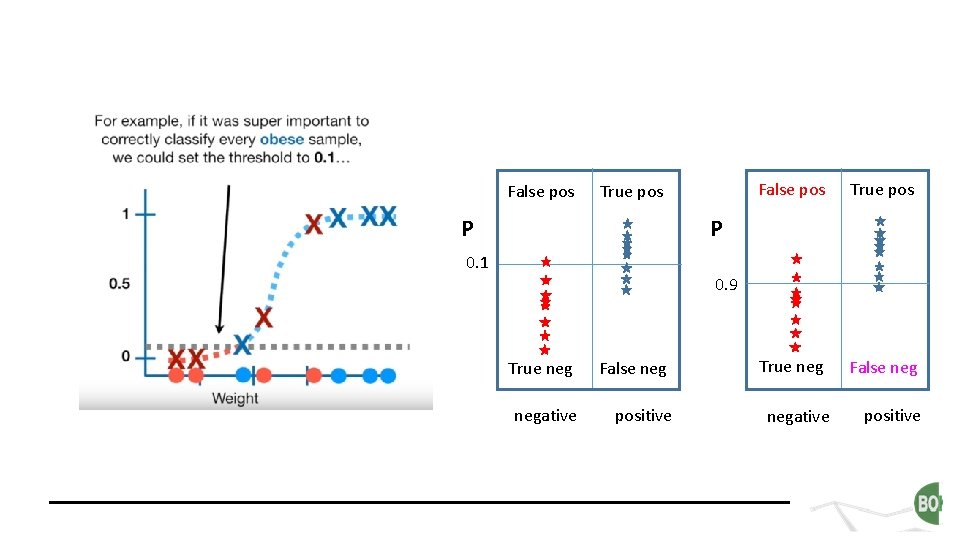
False pos True pos P False pos True neg False neg P 0. 1 0. 9 True neg False negative positive
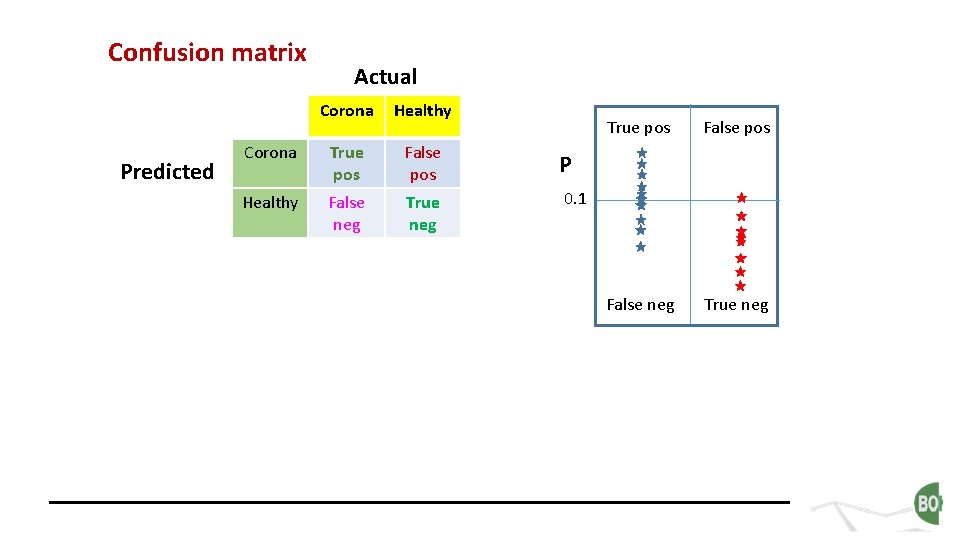
Confusion matrix Predicted Actual Corona Healthy Corona True pos False pos P Healthy False neg True neg 0. 1 True pos False neg True neg
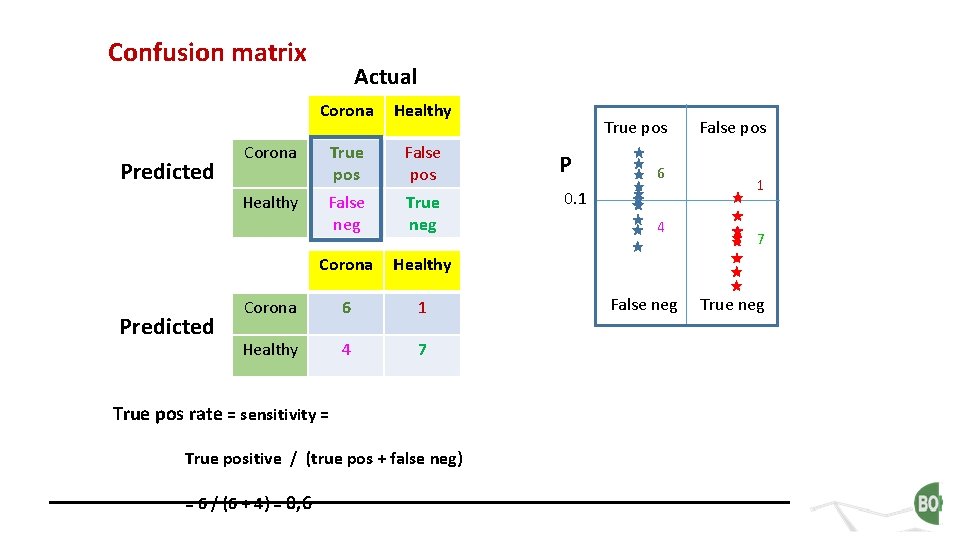
Confusion matrix Predicted Corona Healthy True pos False pos P False neg True neg 0. 1 Corona Healthy Corona 6 1 Healthy 4 7 Corona Healthy Predicted Actual True pos rate = sensitivity = True positive / (true pos + false neg) = 6 / (6 + 4) = 0, 6 True pos 6 4 False neg False pos 1 7 True neg
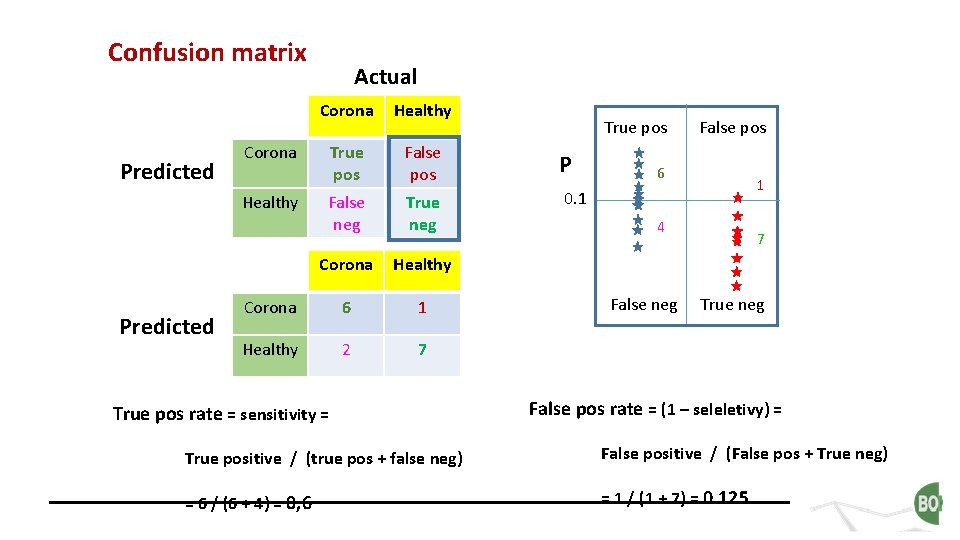
Confusion matrix Predicted Corona Healthy True pos False pos P False neg True neg 0. 1 Corona Healthy Corona 6 1 Healthy 2 7 Corona Healthy Predicted Actual True pos rate = sensitivity = True pos False pos 6 1 4 False neg 7 True neg False pos rate = (1 – seleletivy) = True positive / (true pos + false neg) False positive / (False pos + True neg) = 6 / (6 + 4) = 0, 6 = 1 / (1 + 7) = 0. 125
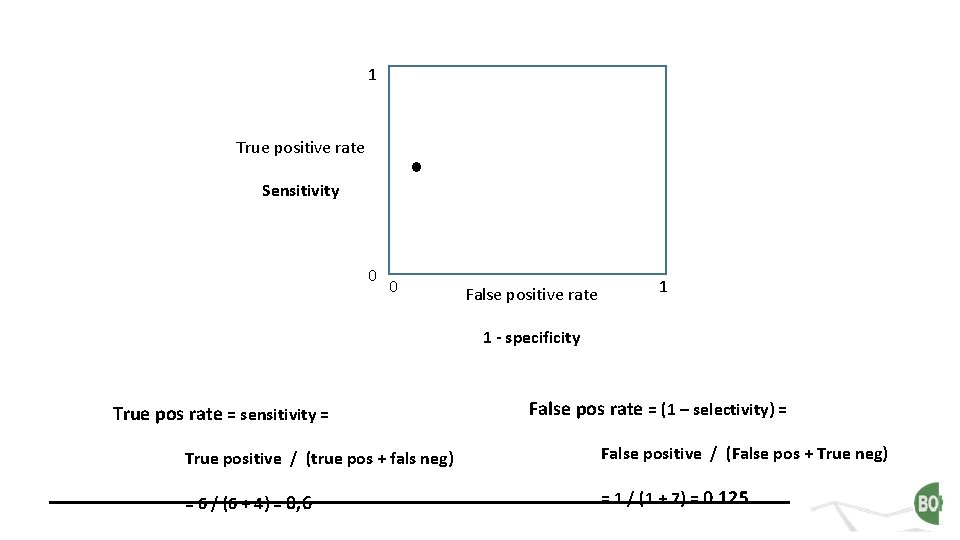
1 True positive rate Sensitivity 0 0 False positive rate 1 1 - specificity True pos rate = sensitivity = False pos rate = (1 – selectivity) = True positive / (true pos + fals neg) False positive / (False pos + True neg) = 6 / (6 + 4) = 0, 6 = 1 / (1 + 7) = 0. 125
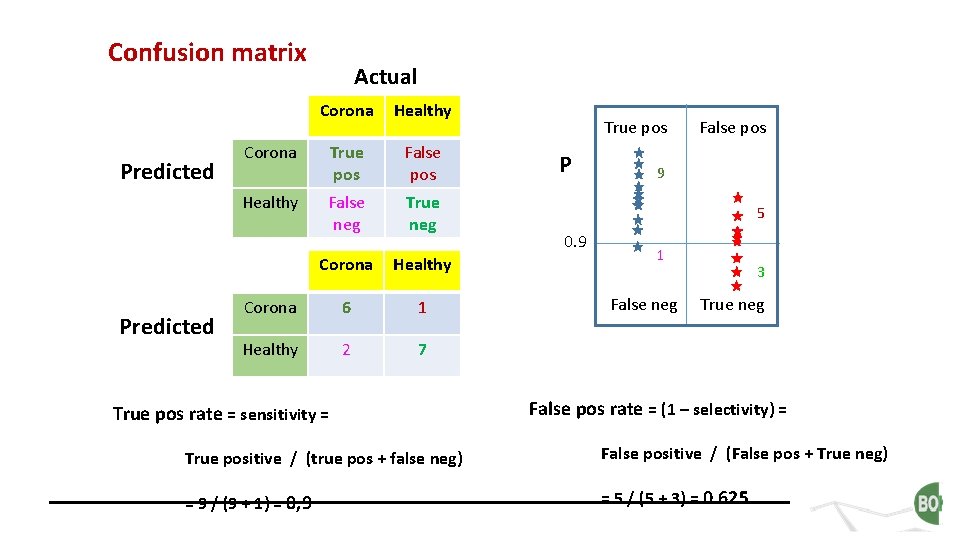
Confusion matrix Predicted Actual Corona Healthy Corona True pos False pos Healthy False neg True neg Corona Healthy Corona 6 1 Healthy 2 7 True pos rate = sensitivity = True pos P False pos 9 5 0. 9 1 False neg 3 True neg False pos rate = (1 – selectivity) = True positive / (true pos + false neg) False positive / (False pos + True neg) = 9 / (9 + 1) = 0, 9 = 5 / (5 + 3) = 0. 625
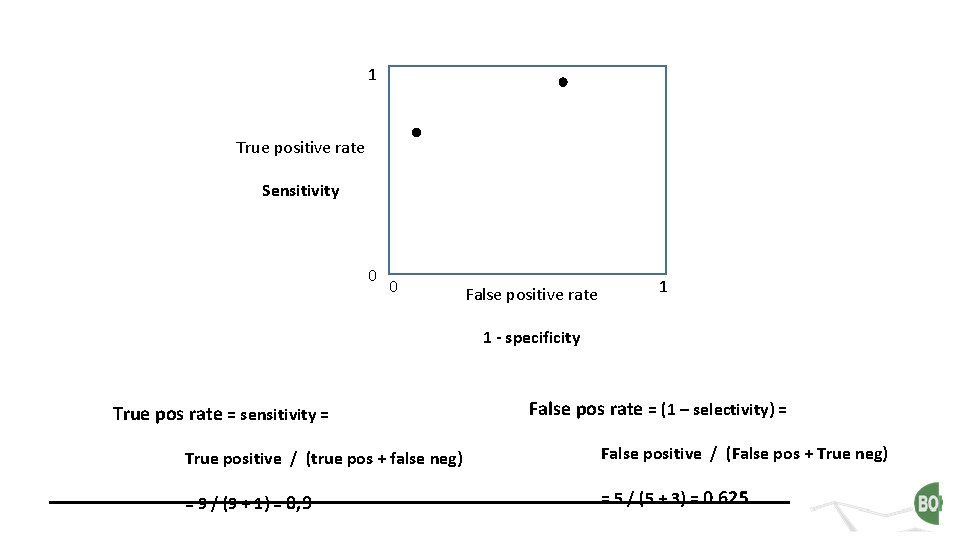
1 True positive rate Sensitivity 0 0 False positive rate 1 1 - specificity True pos rate = sensitivity = False pos rate = (1 – selectivity) = True positive / (true pos + false neg) False positive / (False pos + True neg) = 9 / (9 + 1) = 0, 9 = 5 / (5 + 3) = 0. 625
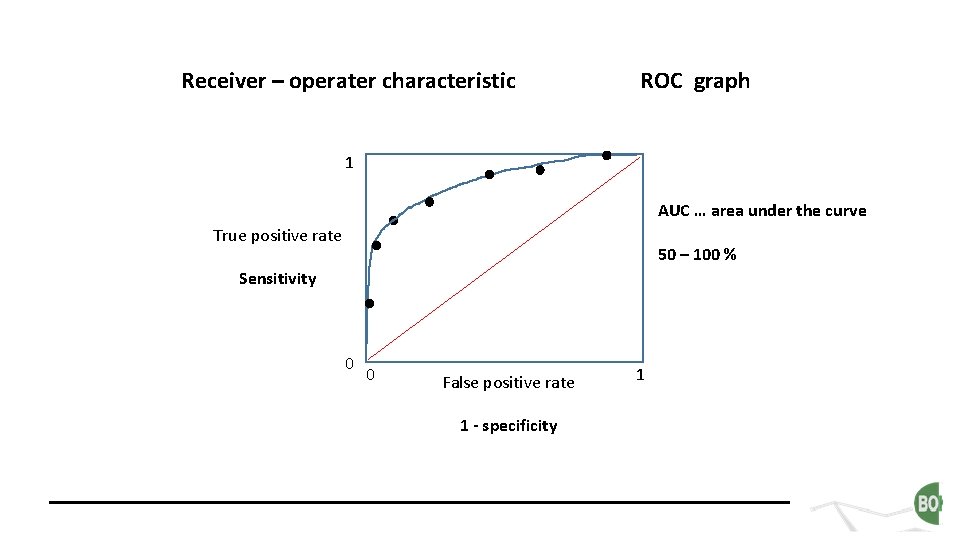
Receiver – operater characteristic ROC graph 1 AUC … area under the curve True positive rate 50 – 100 % Sensitivity 0 0 False positive rate 1 - specificity 1
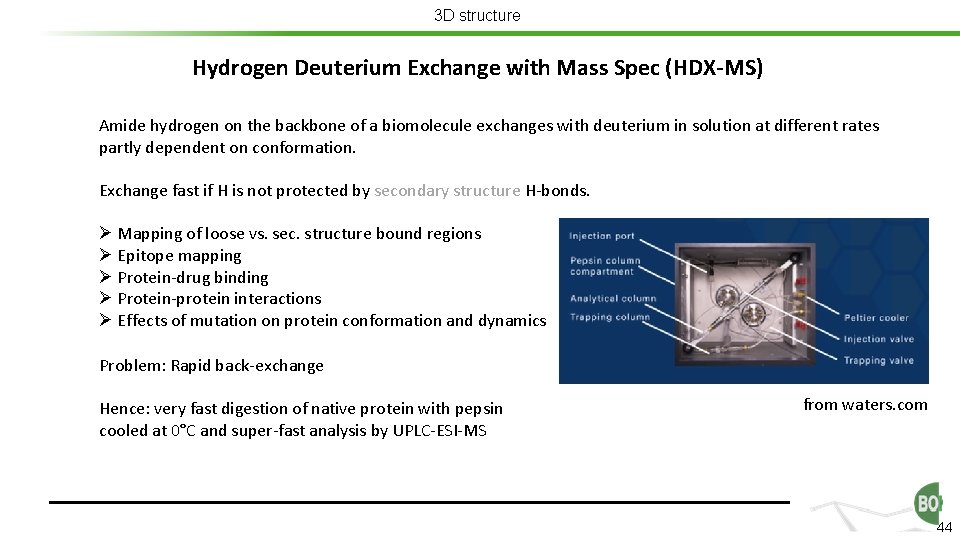
3 D structure Hydrogen Deuterium Exchange with Mass Spec (HDX-MS) Amide hydrogen on the backbone of a biomolecule exchanges with deuterium in solution at different rates partly dependent on conformation. Exchange fast if H is not protected by secondary structure H-bonds. Ø Mapping of loose vs. sec. structure bound regions Ø Epitope mapping Ø Protein-drug binding Ø Protein-protein interactions Ø Effects of mutation on protein conformation and dynamics Problem: Rapid back-exchange Hence: very fast digestion of native protein with pepsin cooled at 0°C and super-fast analysis by UPLC-ESI-MS from waters. com 44
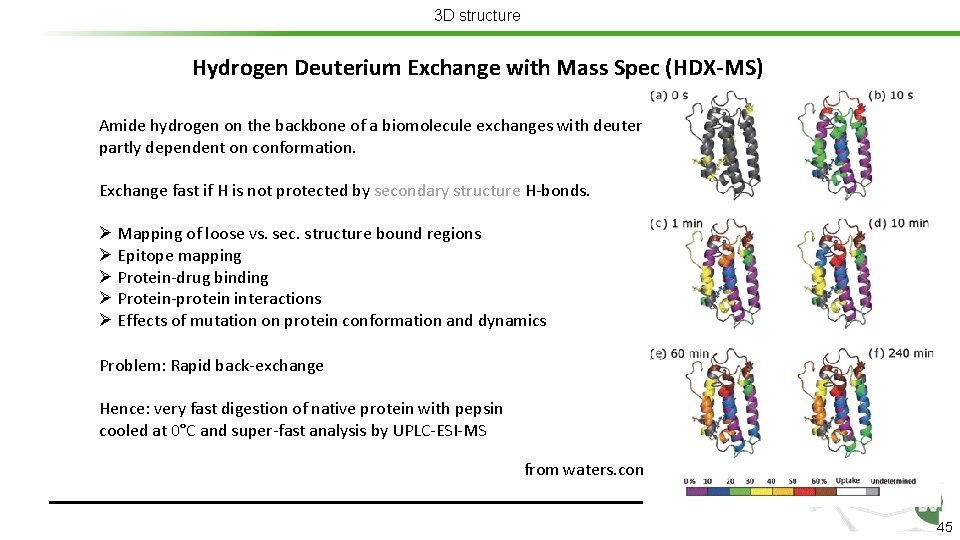
3 D structure Hydrogen Deuterium Exchange with Mass Spec (HDX-MS) Amide hydrogen on the backbone of a biomolecule exchanges with deuterium in solution at different rates partly dependent on conformation. Exchange fast if H is not protected by secondary structure H-bonds. Ø Mapping of loose vs. sec. structure bound regions Ø Epitope mapping Ø Protein-drug binding Ø Protein-protein interactions Ø Effects of mutation on protein conformation and dynamics Problem: Rapid back-exchange Hence: very fast digestion of native protein with pepsin cooled at 0°C and super-fast analysis by UPLC-ESI-MS from waters. com 45

Thank you for listening – its done ! 46
- Slides: 46