Proteomics Analyzing proteins space Protein families Why proteins









- Slides: 25

Proteomics: Analyzing proteins space

Protein families Why proteins? • Shift of interest from “Genomics” to “Proteomics” Classification of proteins to groups/families - what is it good for? • Explosion in biological sequence data => need to organize! • Understanding relations/hierarchy of groups is interesting as is, e. g. in evolutionary research. • For applied research : – Annotation of new proteins : predicting their function, structure, cellular localization etc. – Looking for new folds

Sequence-based classification • By sequence similarity (domains, motifs or complete proteins) : Pfam, PROSITE, SMART, Inter. Pro etc. • Inter. Pro – Synthesizes the data from Pfam, PROSITE, Prints, Pro. Dom, and SMART. Considered as “best” domain-based classification available

Other kinds of classification • Global classification : – Systers, Protomap, CLUSTr – Meta. Fam synthesizes global classification data • By structure similarity : SCOP etc. • By function : Albumin, Ret. Net, Tumor. Genes etc.

http: //www. protonet. cs. huji. ac. il • A long-term project in HUJI led by Michal & Nati Linial. • Provides automatic global classification of the known proteins. • Performs hierarchical clustering on sequence-based metric space of proteins. • Allows to “place” an external protein into the hierarchy.

Why clustering? • • • We want to refine the “similarity” notion, compared to e. g. BLAST Exploit transitivity to improve grouping Can use a low threshold on similarity: - uses vast information from low similarities - allowable because clustering filters noise

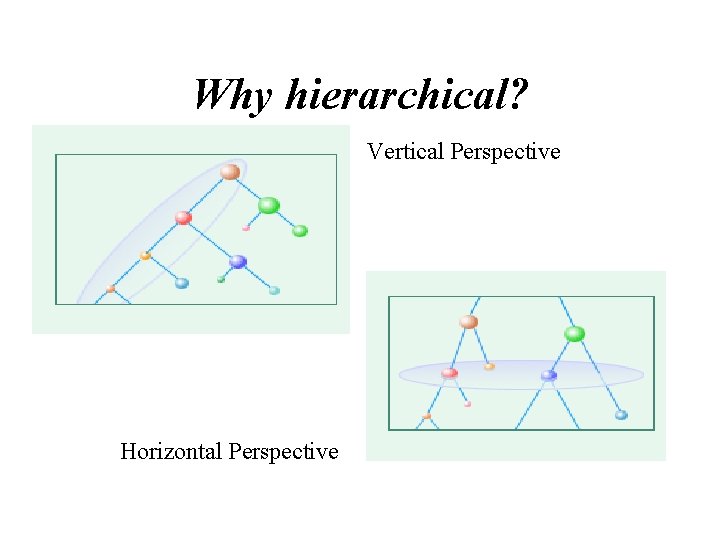
Why hierarchical? Vertical Perspective Horizontal Perspective

Proto. Net: Pre-Computation • All-against-all gapped BLAST using BLOSUM 62 • Swiss. Prot release 40. 28 database (114, 033 proteins) • BLAST identified ~2*107 relations between these proteins with relatively high sequence similarity E-Score of 100 or less: • Don’t want to lose information => very permissive! • But still less then ~6. 5*109 => infeasible

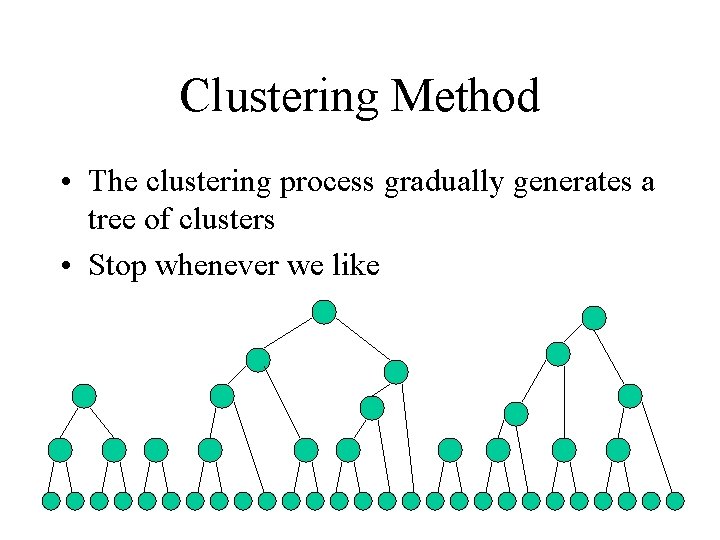
Clustering Method • First, each cluster is considered a singleton

Clustering Method • Next, we iteratively merge the pairs of clusters • We choose to merge the ‘most similar’ pair of clusters.

Clustering Method • Next, we iteratively merge the pairs of clusters • We choose to merge the ‘most similar’ pair of clusters.

Clustering Method • Next, we iteratively merge the pairs of clusters • We choose to merge the ‘most similar’ pair of clusters.

Clustering Method • As we progress the number of singletons drops

Clustering Method • The clustering process gradually generates a tree of clusters • Stop whenever we like

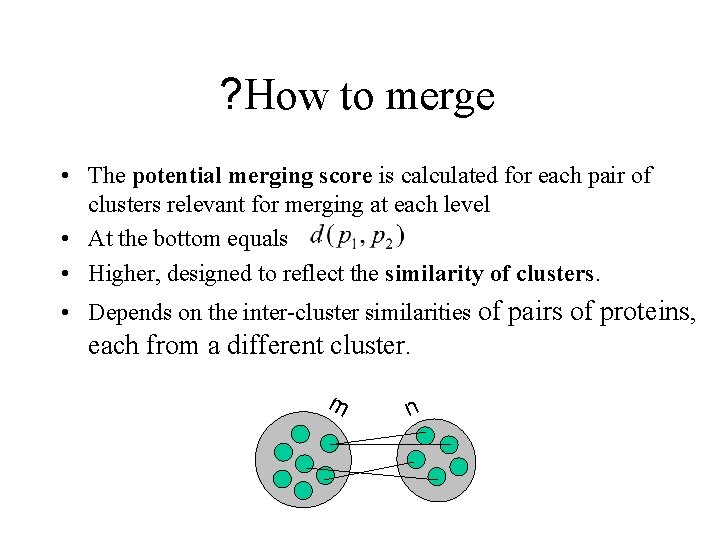
? How to merge • The potential merging score is calculated for each pair of clusters relevant for merging at each level • At the bottom equals • Higher, designed to reflect the similarity of clusters. • Depends on the inter-cluster similarities of pairs of proteins, each from a different cluster. m n

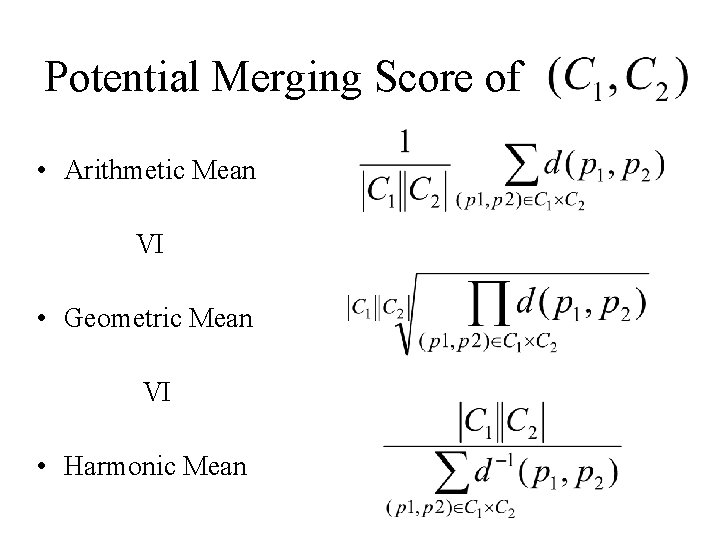
Potential Merging Score of • Arithmetic Mean VI • Geometric Mean VI • Harmonic Mean

Missing Data Treatment • For very low similarity pair (outside of ~2*107 ), its length is defined as • Practically, the merging process should finish, when the weight of the “infinite” lengths in calculation of the score between new clusters is very large (losing signal)

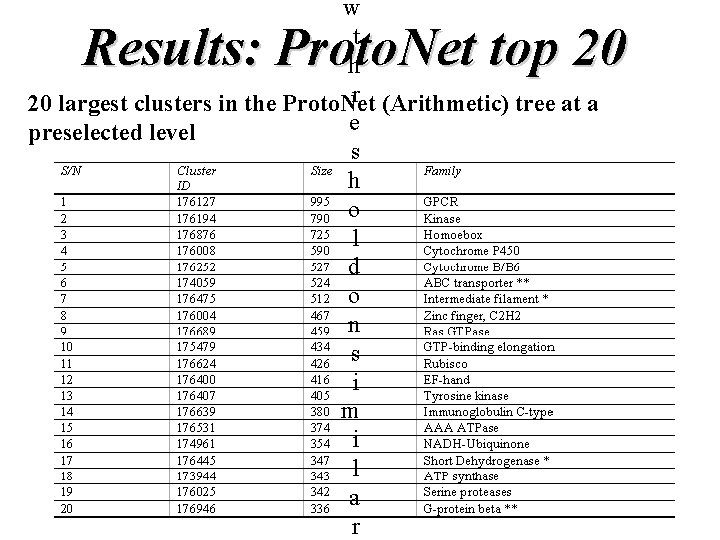
w t h r (Arithmetic) tree at a 20 largest clusters in the Proto. Net e preselected level s h o l d o n s i m i l a r Results: Proto. Net top 20

Problem of result assessment: what is a “good” cluster? • Contains all proteins in the family, does not contain proteins not in family • But what is family? Does any keyword define a family? • Stable as the merging events occur (long lifetime)?

Problem of result assessment: what is a “good” tree? • Should we trust the resulting forest? – Which clustering technique is better? Combined? – Bootstrap? • Do the clusters correspond to meaningful families of proteins? – Validation against Inter. Pro, SCOP etc. – Lack of will to automatically reconstruct them!!! • What is the right level/cut to look at the forest?

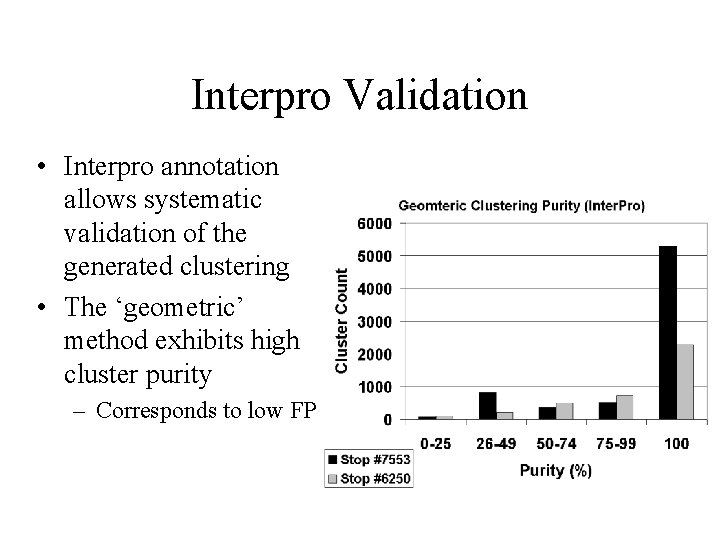
Interpro Validation • Interpro annotation allows systematic validation of the generated clustering • The ‘geometric’ method exhibits high cluster purity – Corresponds to low FP

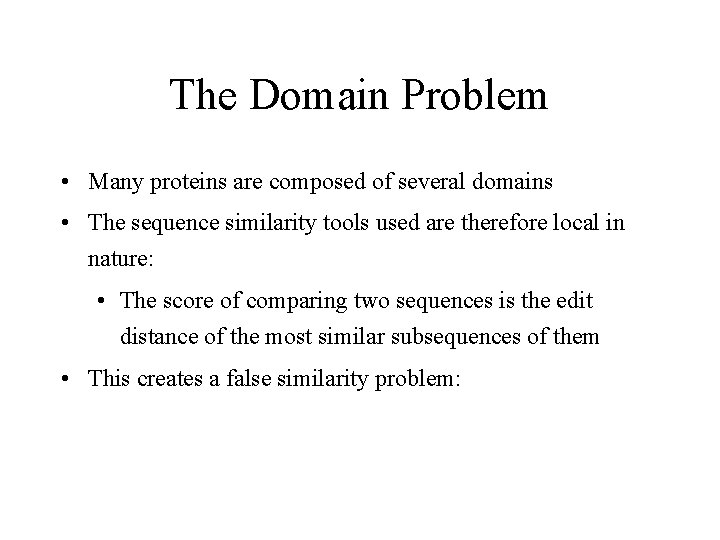
The Domain Problem • Many proteins are composed of several domains • The sequence similarity tools used are therefore local in nature: • The score of comparing two sequences is the edit distance of the most similar subsequences of them • This creates a false similarity problem:

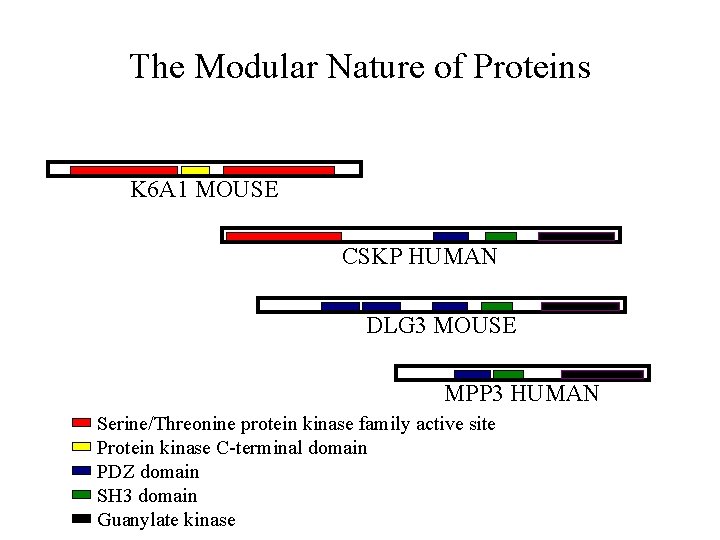
The Modular Nature of Proteins K 6 A 1 MOUSE CSKP HUMAN DLG 3 MOUSE MPP 3 HUMAN Serine/Threonine protein kinase family active site Protein kinase C-terminal domain PDZ domain SH 3 domain Guanylate kinase

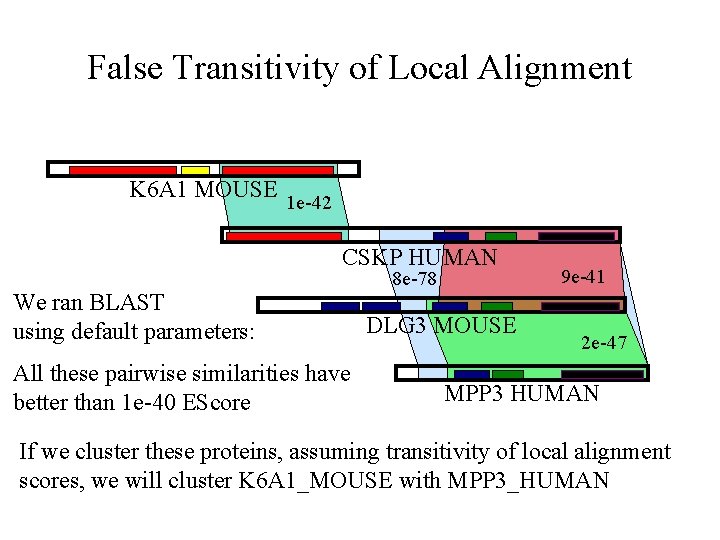
False Transitivity of Local Alignment K 6 A 1 MOUSE 1 e-42 CSKP HUMAN We ran BLAST using default parameters: All these pairwise similarities have better than 1 e-40 EScore 8 e-78 DLG 3 MOUSE 9 e-41 2 e-47 MPP 3 HUMAN If we cluster these proteins, assuming transitivity of local alignment scores, we will cluster K 6 A 1_MOUSE with MPP 3_HUMAN

Alternative methods • Different types of clustering – Non-binary – Goal-oriented => semi-guided – Graph theory insights • Non-clustering ways of exploring the space of proteins • Why BLAST E-score? ? ? • Enrichment of the metric using structure