Protein Transport into the Mitochondria and Chloroplast Subcompartments

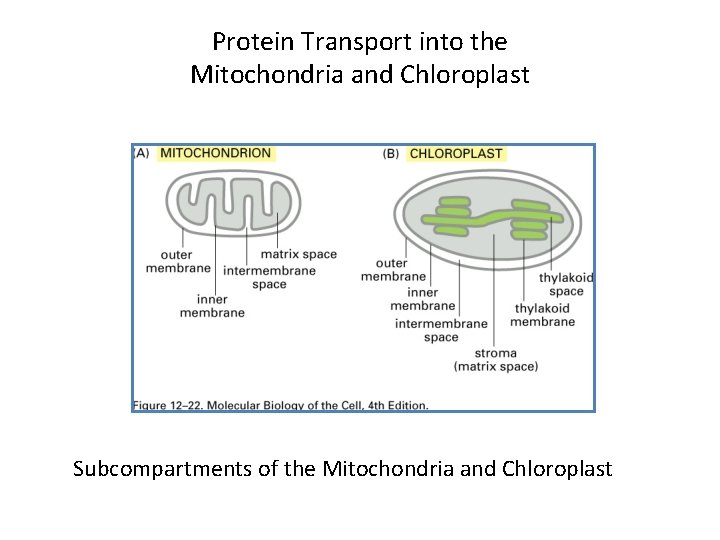
Protein Transport into the Mitochondria and Chloroplast Subcompartments of the Mitochondria and Chloroplast
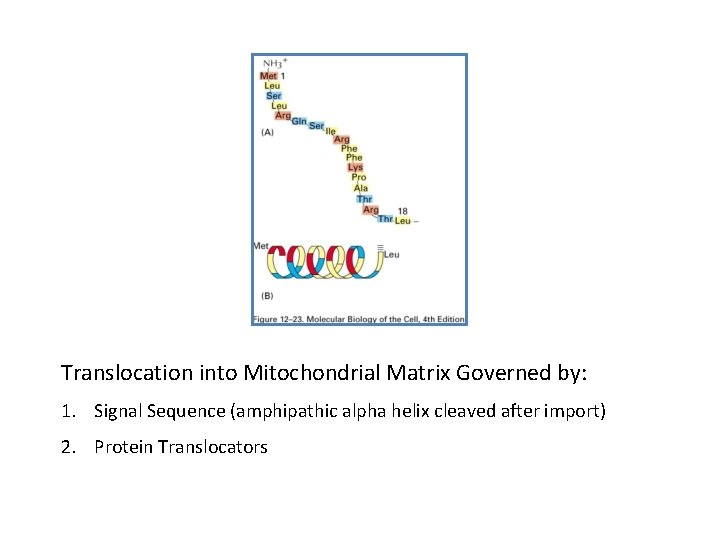
Translocation into Mitochondrial Matrix Governed by: 1. Signal Sequence (amphipathic alpha helix cleaved after import) 2. Protein Translocators
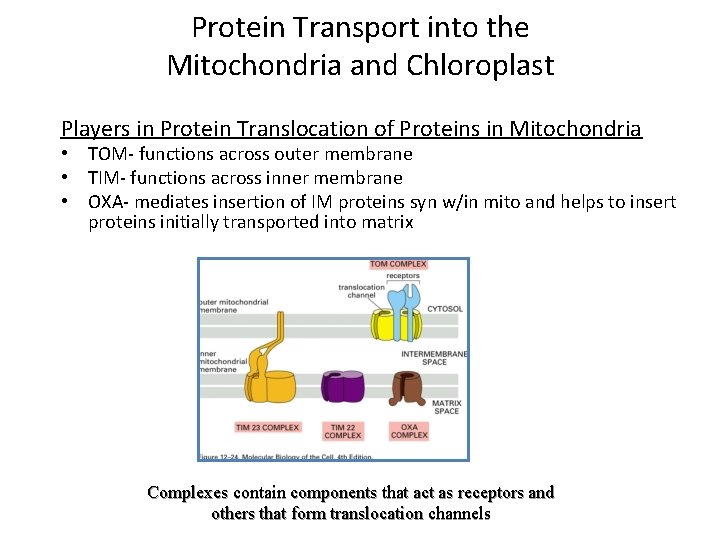
Protein Transport into the Mitochondria and Chloroplast Players in Protein Translocation of Proteins in Mitochondria • TOM- functions across outer membrane • TIM- functions across inner membrane • OXA- mediates insertion of IM proteins syn w/in mito and helps to insert proteins initially transported into matrix Complexes contain components that act as receptors and others that form translocation channels
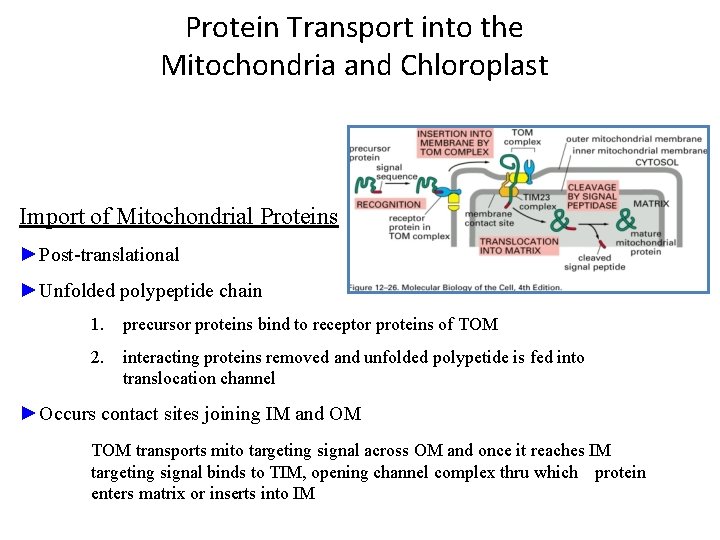
Protein Transport into the Mitochondria and Chloroplast Import of Mitochondrial Proteins ►Post-translational ►Unfolded polypeptide chain 1. precursor proteins bind to receptor proteins of TOM 2. interacting proteins removed and unfolded polypetide is fed into translocation channel ►Occurs contact sites joining IM and OM TOM transports mito targeting signal across OM and once it reaches IM targeting signal binds to TIM, opening channel complex thru which protein enters matrix or inserts into IM
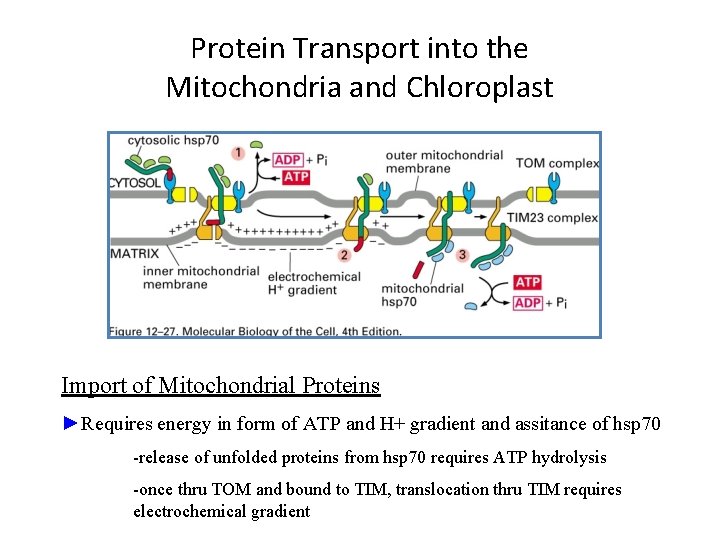
Protein Transport into the Mitochondria and Chloroplast Import of Mitochondrial Proteins ►Requires energy in form of ATP and H+ gradient and assitance of hsp 70 -release of unfolded proteins from hsp 70 requires ATP hydrolysis -once thru TOM and bound to TIM, translocation thru TIM requires electrochemical gradient
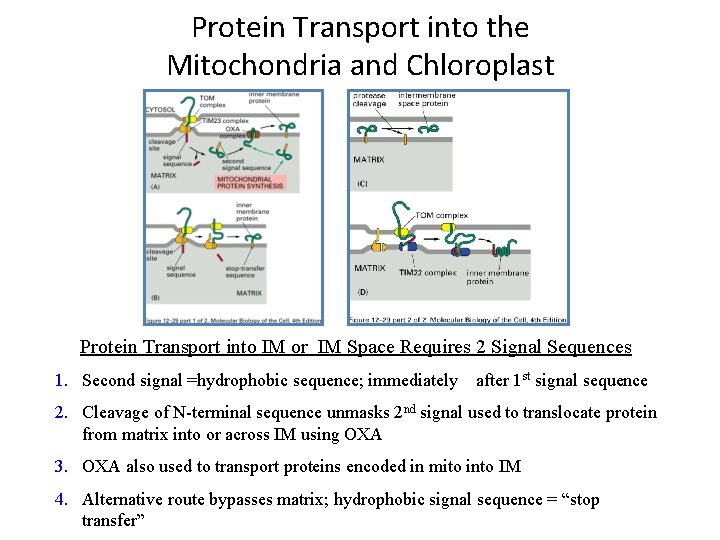
Protein Transport into the Mitochondria and Chloroplast Protein Transport into IM or IM Space Requires 2 Signal Sequences 1. Second signal =hydrophobic sequence; immediately after 1 st signal sequence 2. Cleavage of N-terminal sequence unmasks 2 nd signal used to translocate protein from matrix into or across IM using OXA 3. OXA also used to transport proteins encoded in mito into IM 4. Alternative route bypasses matrix; hydrophobic signal sequence = “stop transfer”
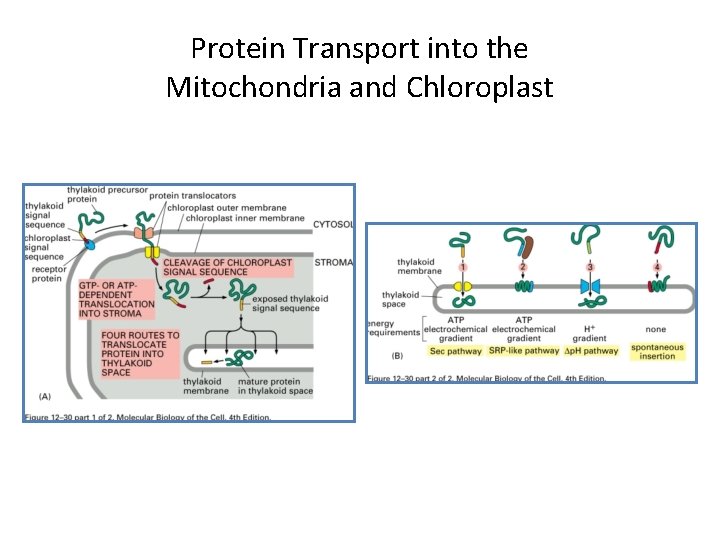
Protein Transport into the Mitochondria and Chloroplast Protein Transport into Chloro Similar to Transport into Mito 1. 2. 3. 4. 5. 6. occur posttranslationally Use separate translocation complexes in ea membrane Translocation occurs at contact sites Requires energy and electrochemical gradient Use amphilpathic N-terminal signal seq that is removed Like the mito a second signal sequence required for translocation into thylakoid mem or space
Protein Transport into the Mitochondria and Chloroplast
Peroxisomes and Protein Import Peroxisomes ►Use O 2 and H 2 O 2 to carry out oxidative rxns ►Remove H from specific organic compounds RH 2 + O 2 R + H 2 O 2 ►Catalases use H 2 O 2 to oxidize other substances, particularly in liver and kidney detoxification H 2 O 2 + R’H 2 R’ + H 2 O ►Beta Oxidation ►Formation of plasmalogens (abundant class of phospholipids in myelin) ►Photorespiration and glyoxylate cycle in plants

b OSSIDAZIONE DEGLI ACIDI GRASSI - Reazione di demolizione degli acidi grassi: Le catene alchiliche sono accorciate in sequenze di 2 atomi di carbonio alla volta (acetil Co. A) - Acetil Co. A è esportato al citolsol dove viene usato nelle reazioni biosintetiche - reazione effettuata anche nei mitocondri nelle cellule di mammifero

I PEROSSISOMI Organelli a singola membrana presenti in tutte le cellule - Contengono enzimi ossidativi come la CATALASI -Rappresentano il sito di utilizzo dell’ossigeno molecolare -Le proteine dei perossisomi derivano da ribosomi liberi -Si pensa che siano vestigia di un antico organello che svolgeva il metabolismo dell’ossigeno negli antenati delle cellule eucariotiche. Gli enzimi utilizzano l’ossigeno molecolare per rimuovere atomi di H da substrati organici in reazioni ossidative che producono perossido di idrogeno.
Peroxisomes and Protein Import Peroximsomes in Plants ►Site of Photorespiration= glycolate pathway in leaves ►Called glyoxysomes in seeds where fats converted into sugar
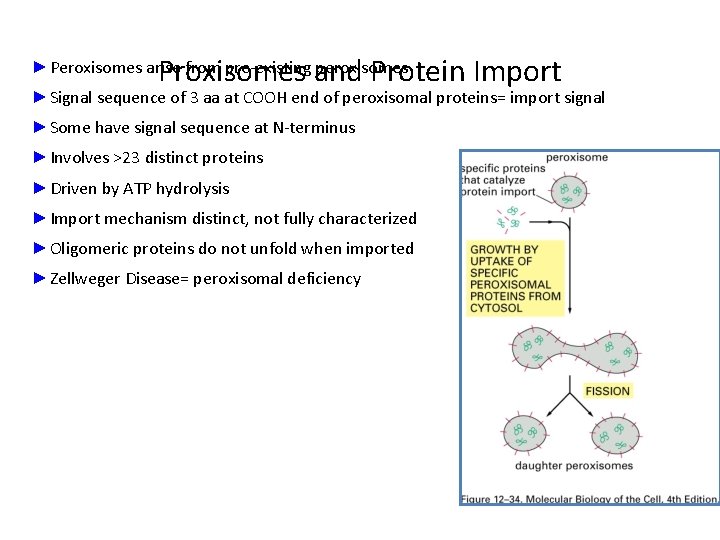
Proxisomes and Protein Import ►Peroxisomes arise from pre-existing peroxisomes ►Signal sequence of 3 aa at COOH end of peroxisomal proteins= import signal ►Some have signal sequence at N-terminus ►Involves >23 distinct proteins ►Driven by ATP hydrolysis ►Import mechanism distinct, not fully characterized ►Oligomeric proteins do not unfold when imported ►Zellweger Disease= peroxisomal deficiency
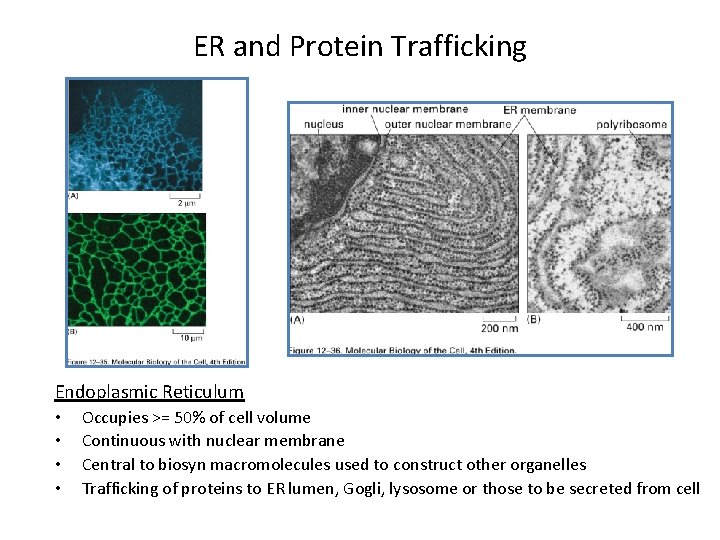
ER and Protein Trafficking Endoplasmic Reticulum • • Occupies >= 50% of cell volume Continuous with nuclear membrane Central to biosyn macromolecules used to construct other organelles Trafficking of proteins to ER lumen, Gogli, lysosome or those to be secreted from cell
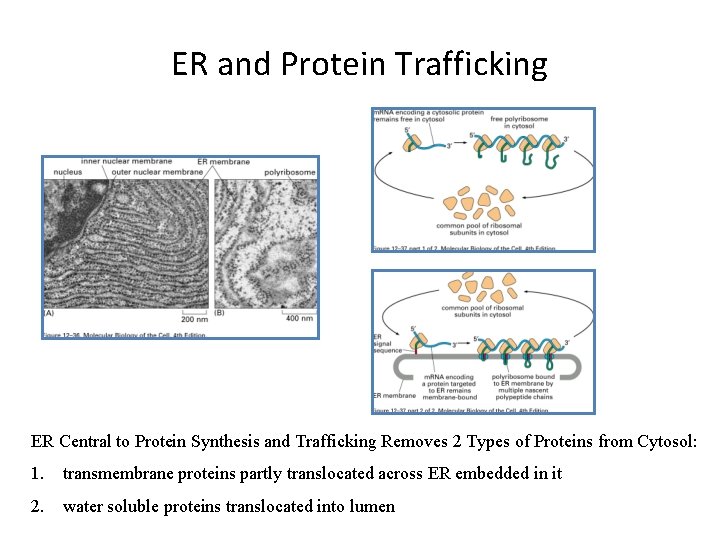
ER and Protein Trafficking ER Central to Protein Synthesis and Trafficking Removes 2 Types of Proteins from Cytosol: 1. transmembrane proteins partly translocated across ER embedded in it 2. water soluble proteins translocated into lumen
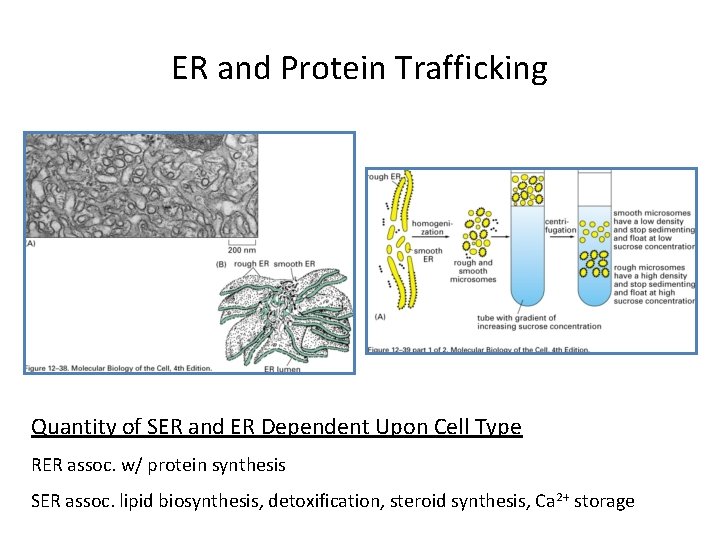
ER and Protein Trafficking Quantity of SER and ER Dependent Upon Cell Type RER assoc. w/ protein synthesis SER assoc. lipid biosynthesis, detoxification, steroid synthesis, Ca 2+ storage
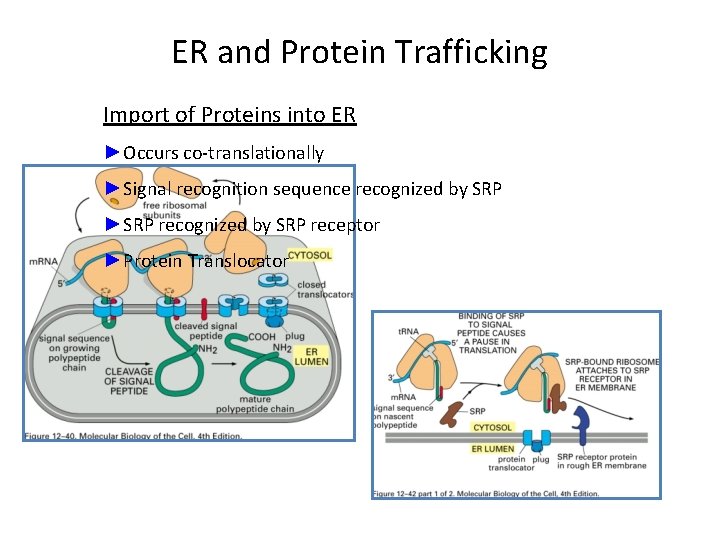
ER and Protein Trafficking Import of Proteins into ER ►Occurs co-translationally ►Signal recognition sequence recognized by SRP ►SRP recognized by SRP receptor ►Protein Translocator
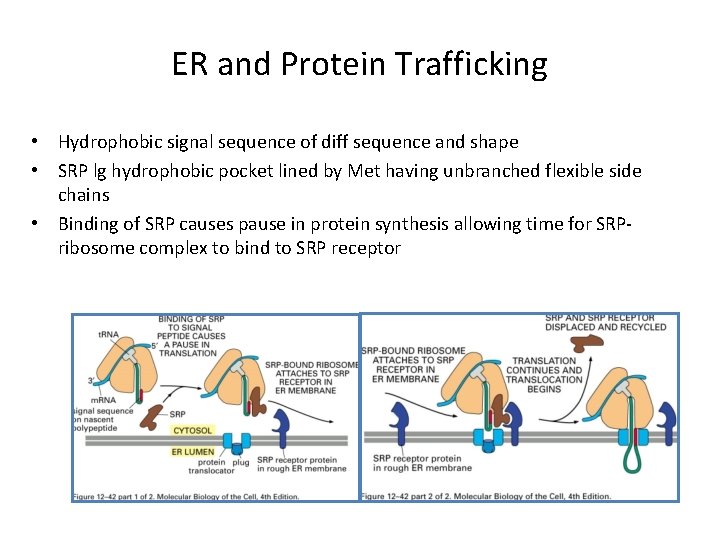
ER and Protein Trafficking • Hydrophobic signal sequence of diff sequence and shape • SRP lg hydrophobic pocket lined by Met having unbranched flexible side chains • Binding of SRP causes pause in protein synthesis allowing time for SRPribosome complex to bind to SRP receptor
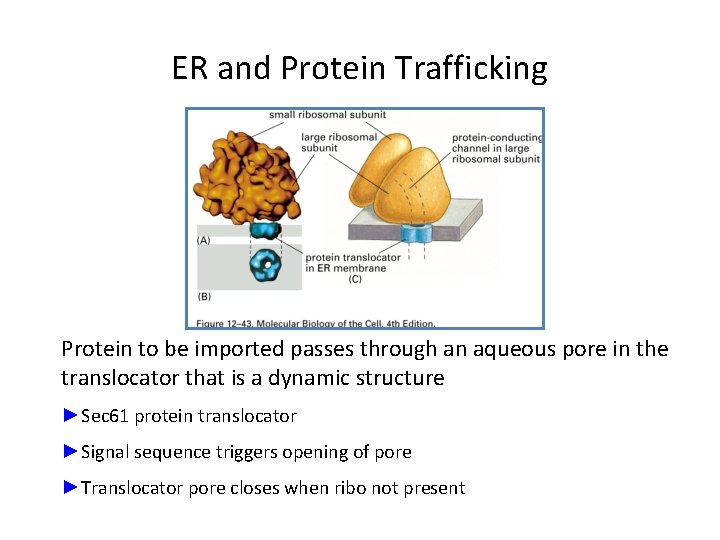
ER and Protein Trafficking Protein to be imported passes through an aqueous pore in the translocator that is a dynamic structure ►Sec 61 protein translocator ►Signal sequence triggers opening of pore ►Translocator pore closes when ribo not present
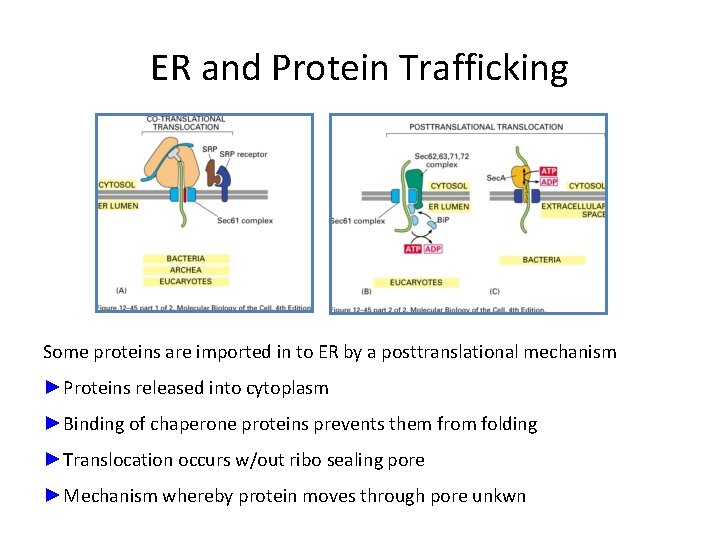
ER and Protein Trafficking Some proteins are imported in to ER by a posttranslational mechanism ►Proteins released into cytoplasm ►Binding of chaperone proteins prevents them from folding ►Translocation occurs w/out ribo sealing pore ►Mechanism whereby protein moves through pore unkwn
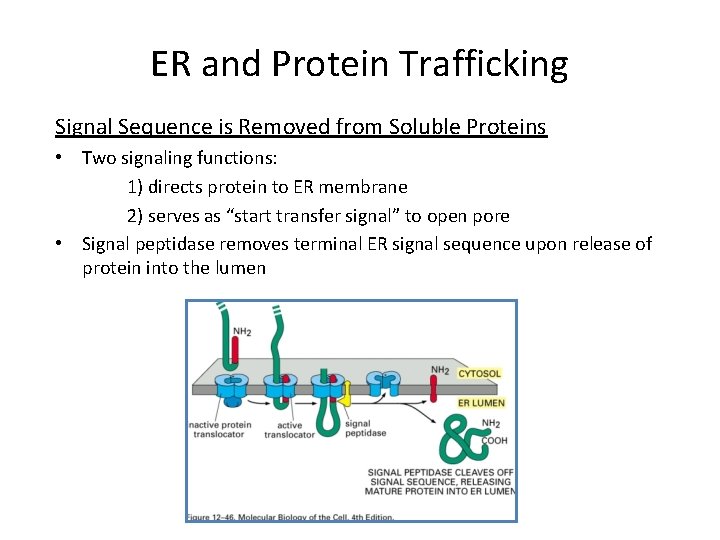
ER and Protein Trafficking Signal Sequence is Removed from Soluble Proteins • Two signaling functions: 1) directs protein to ER membrane 2) serves as “start transfer signal” to open pore • Signal peptidase removes terminal ER signal sequence upon release of protein into the lumen
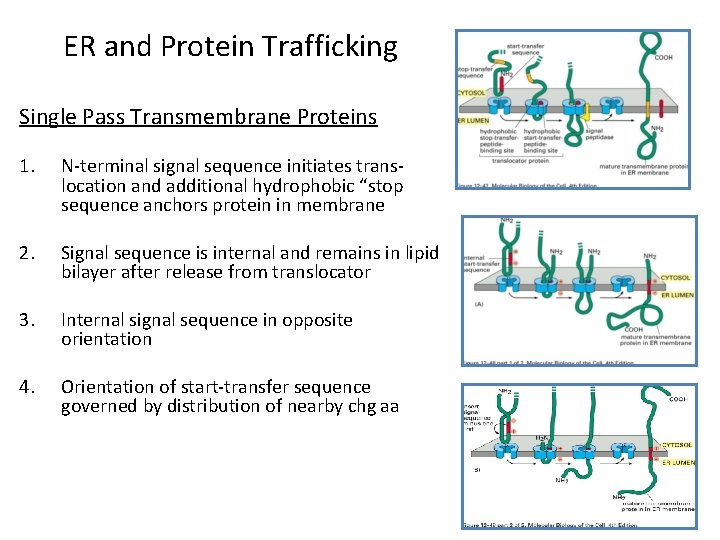
ER and Protein Trafficking Single Pass Transmembrane Proteins 1. N-terminal signal sequence initiates translocation and additional hydrophobic “stop sequence anchors protein in membrane 2. Signal sequence is internal and remains in lipid bilayer after release from translocator 3. Internal signal sequence in opposite orientation 4. Orientation of start-transfer sequence governed by distribution of nearby chg aa
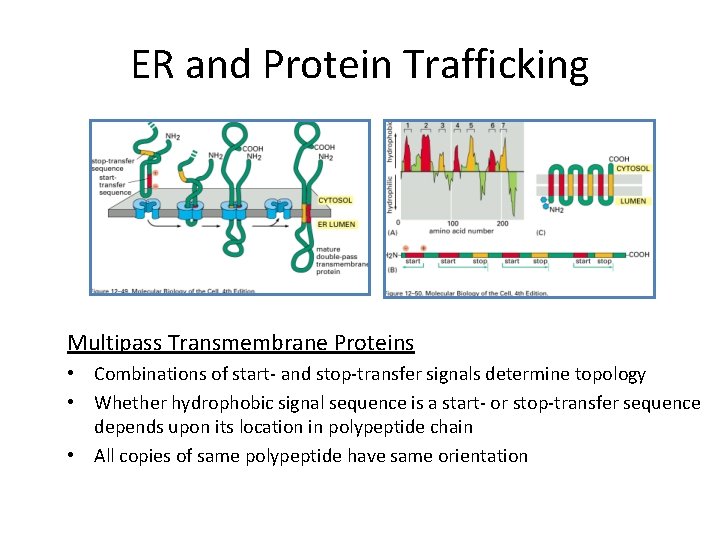
ER and Protein Trafficking Multipass Transmembrane Proteins • Combinations of start- and stop-transfer signals determine topology • Whether hydrophobic signal sequence is a start- or stop-transfer sequence depends upon its location in polypeptide chain • All copies of same polypeptide have same orientation
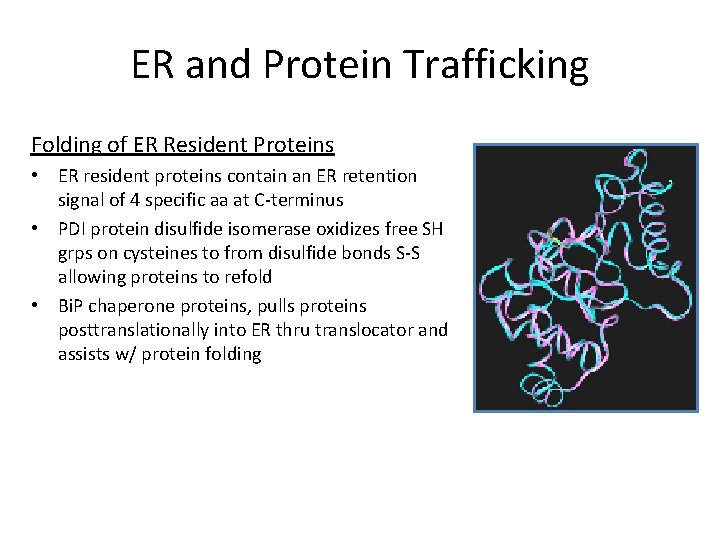
ER and Protein Trafficking Folding of ER Resident Proteins • ER resident proteins contain an ER retention signal of 4 specific aa at C-terminus • PDI protein disulfide isomerase oxidizes free SH grps on cysteines to from disulfide bonds S-S allowing proteins to refold • Bi. P chaperone proteins, pulls proteins posttranslationally into ER thru translocator and assists w/ protein folding
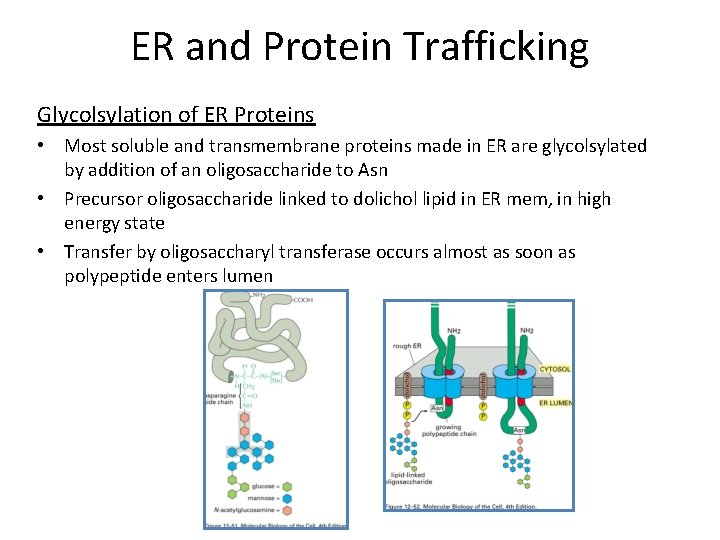
ER and Protein Trafficking Glycolsylation of ER Proteins • Most soluble and transmembrane proteins made in ER are glycolsylated by addition of an oligosaccharide to Asn • Precursor oligosaccharide linked to dolichol lipid in ER mem, in high energy state • Transfer by oligosaccharyl transferase occurs almost as soon as polypeptide enters lumen
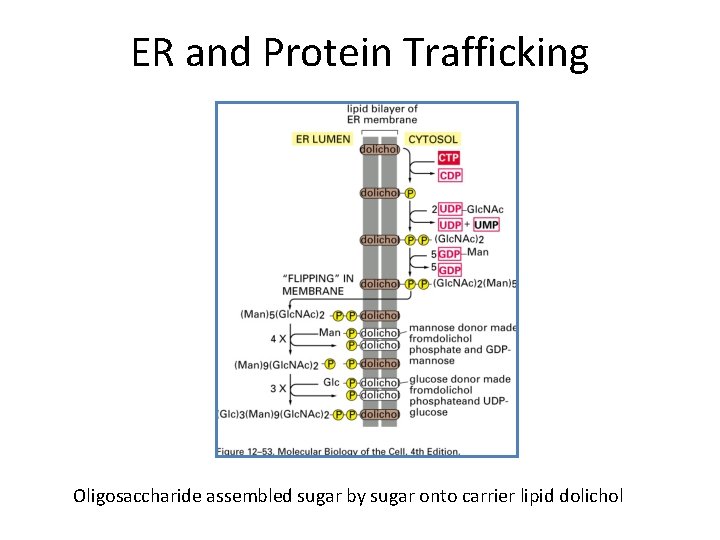
ER and Protein Trafficking Oligosaccharide assembled sugar by sugar onto carrier lipid dolichol
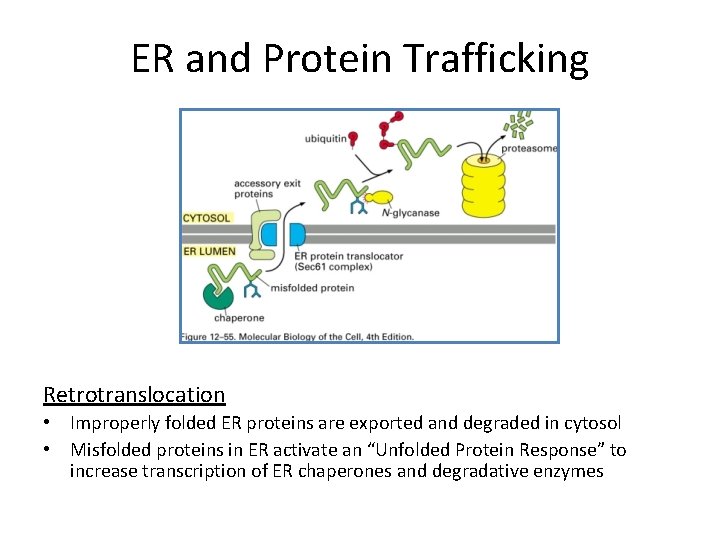
ER and Protein Trafficking Retrotranslocation • Improperly folded ER proteins are exported and degraded in cytosol • Misfolded proteins in ER activate an “Unfolded Protein Response” to increase transcription of ER chaperones and degradative enzymes
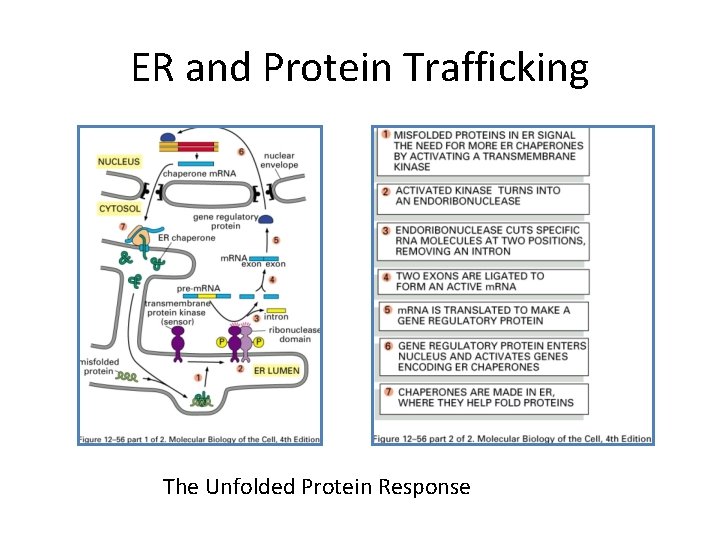
ER and Protein Trafficking The Unfolded Protein Response
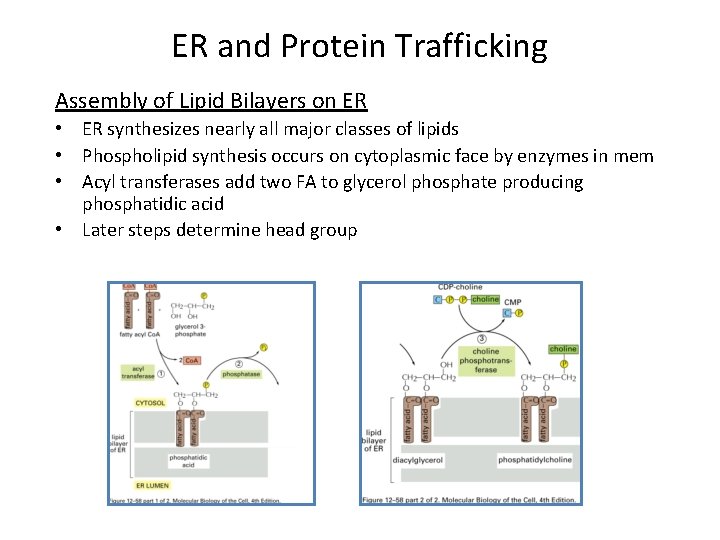
ER and Protein Trafficking Assembly of Lipid Bilayers on ER • ER synthesizes nearly all major classes of lipids • Phospholipid synthesis occurs on cytoplasmic face by enzymes in mem • Acyl transferases add two FA to glycerol phosphate producing phosphatidic acid • Later steps determine head group
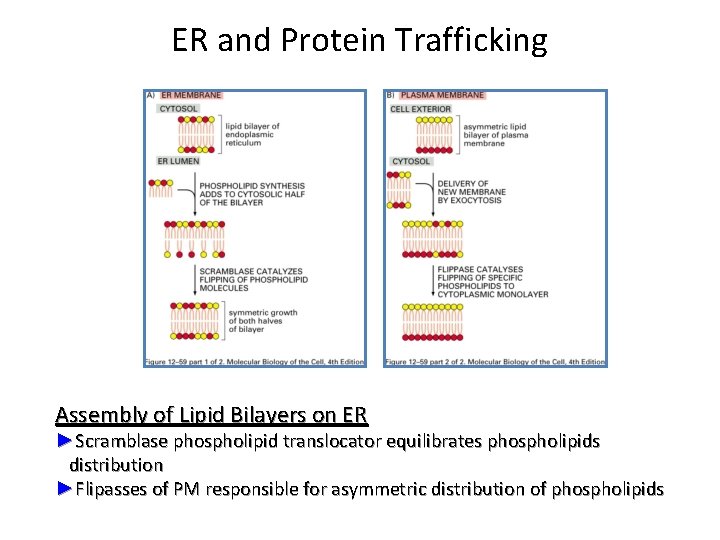
ER and Protein Trafficking Assembly of Lipid Bilayers on ER ►Scramblase phospholipid translocator equilibrates phospholipids distribution ►Flipasses of PM responsible for asymmetric distribution of phospholipids
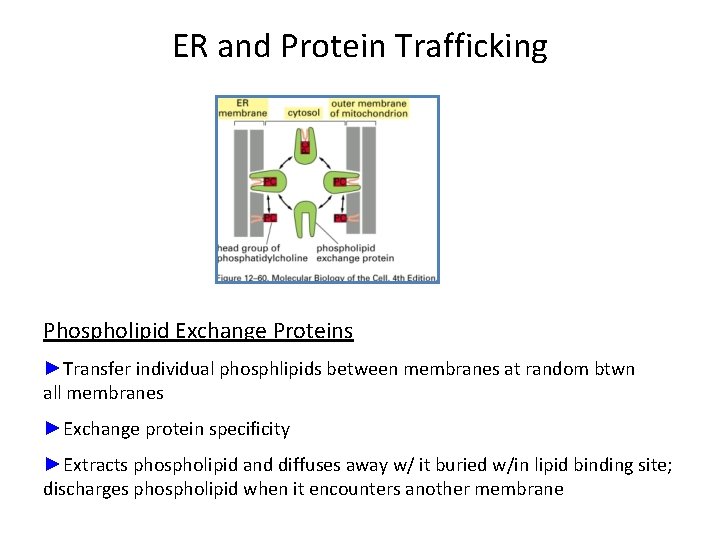
ER and Protein Trafficking Phospholipid Exchange Proteins ►Transfer individual phosphlipids between membranes at random btwn all membranes ►Exchange protein specificity ►Extracts phospholipid and diffuses away w/ it buried w/in lipid binding site; discharges phospholipid when it encounters another membrane

Transport of Molecules Btwn Nucleus and Cytosol
- Slides: 33