PROTEIN SYNTHESIS Big Picture Make a copy of
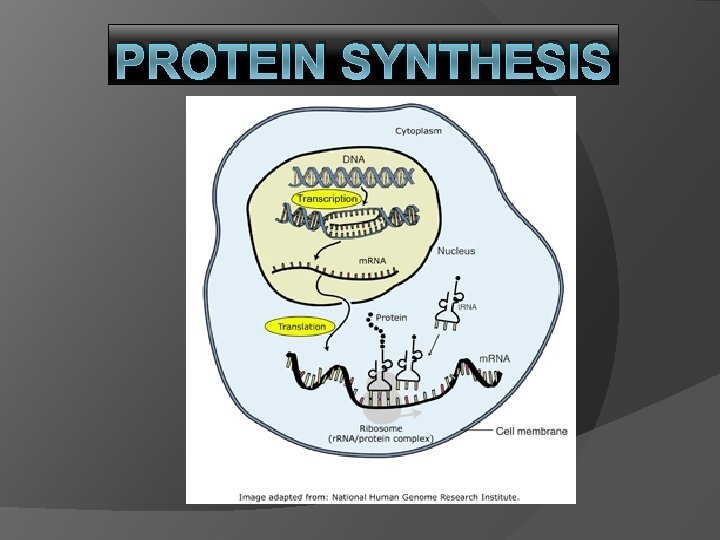
PROTEIN SYNTHESIS

Big Picture Make a copy of DNA (in nucleus) Send copy out of nucleus into cytoplasm Read copy on ribosome Make a protein “Central Dogma”

Organelles and cell machinery involved in protein synthesis Nucleus & Nucleolus Ribosomes Endoplasmic Reticulum Golgi Apparatus Vesicles
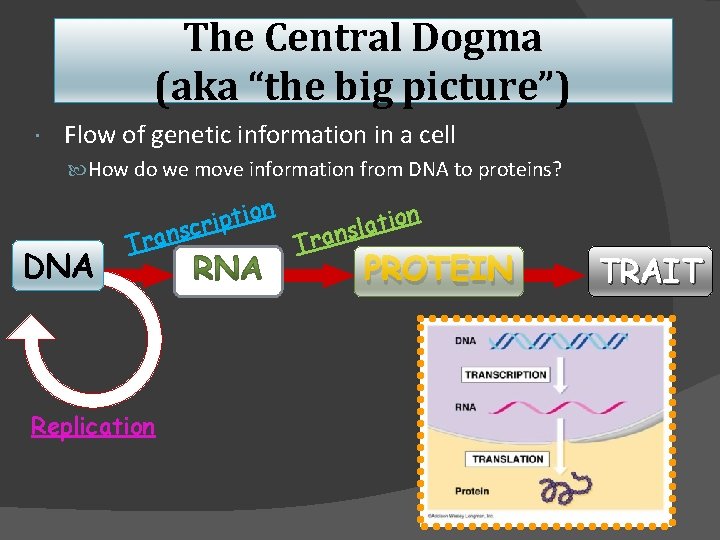
The Central Dogma (aka “the big picture”) Flow of genetic information in a cell How do we move information from DNA to proteins? DNA Tran Replication on i t p i scr on i t a l s n a r T PROTEIN TRAIT

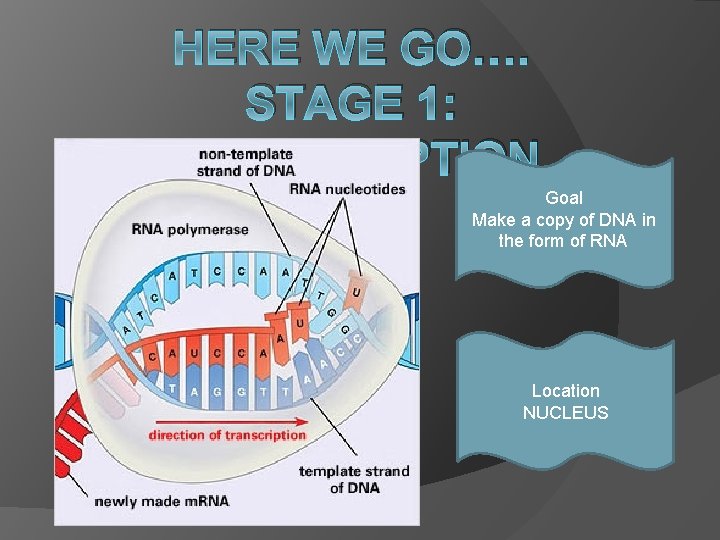
HERE WE GO…. STAGE 1: TRANSCRIPTION Goal Make a copy of DNA in the form of RNA Location NUCLEUS
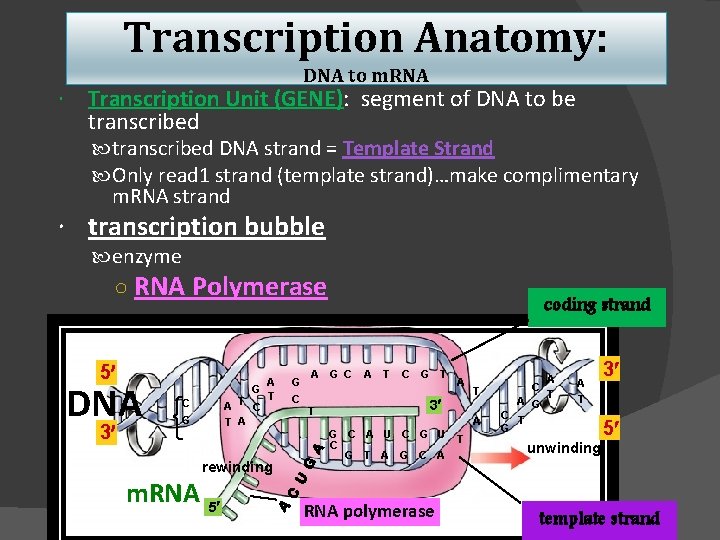
Transcription Anatomy: DNA to m. RNA Transcription Unit (GENE): segment of DNA to be transcribed DNA strand = Template Strand Only read 1 strand (template strand)…make complimentary m. RNA strand transcription bubble enzyme ○ RNA Polymerase 5 DNA 3 A G T A T C T A C G rewinding m. RNA 5 G C coding strand A G C T A T C G T A 3 T A G C A U C G T A G C A RNA polymerase T C A G C T G A T unwinding 3 5 template strand
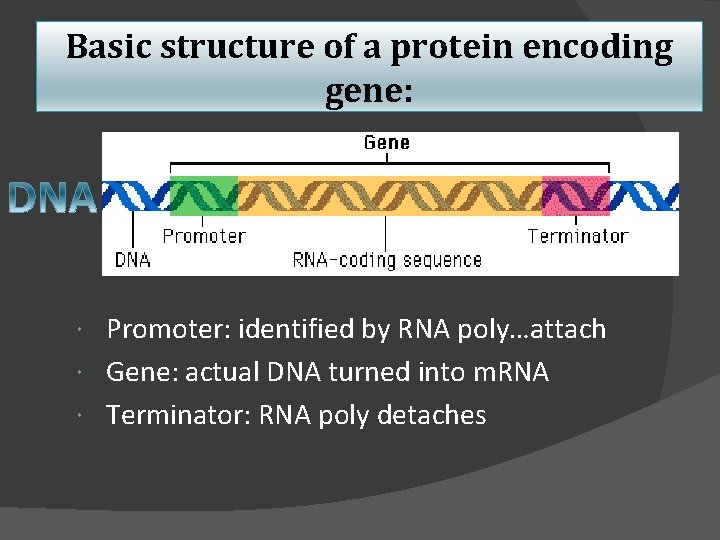
Basic structure of a protein encoding gene: Promoter: identified by RNA poly…attach Gene: actual DNA turned into m. RNA Terminator: RNA poly detaches
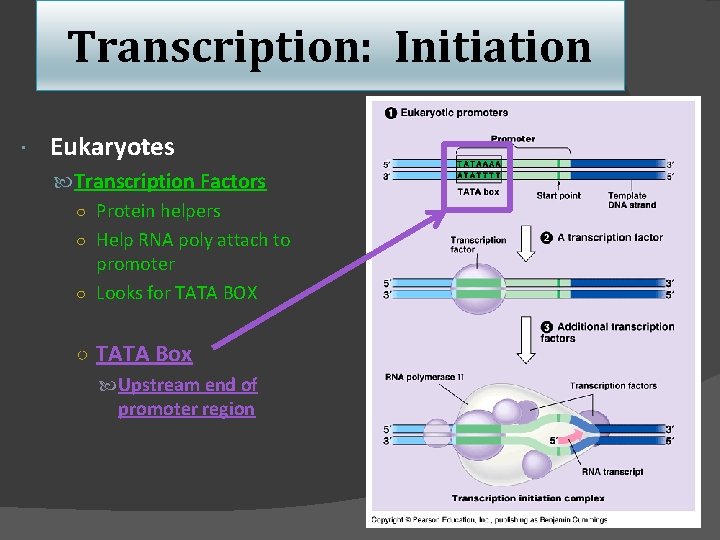
Transcription: Initiation Eukaryotes Transcription Factors ○ Protein helpers ○ Help RNA poly attach to promoter ○ Looks for TATA BOX ○ TATA Box Upstream end of promoter region
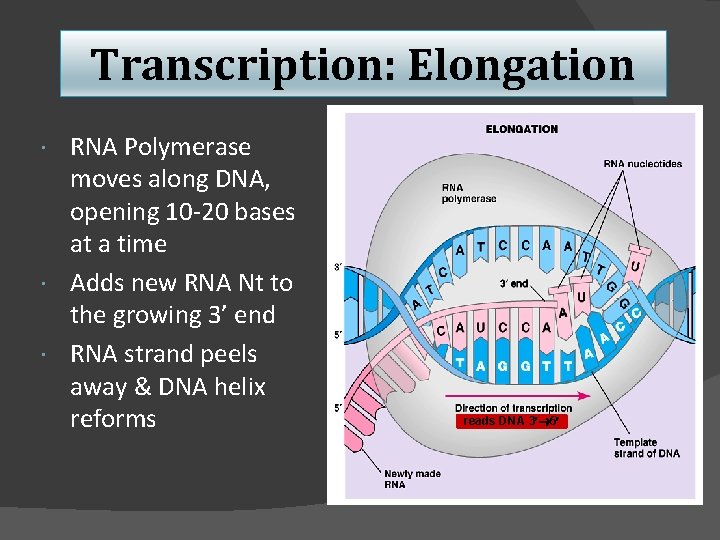
Transcription: Elongation RNA Polymerase moves along DNA, opening 10 -20 bases at a time Adds new RNA Nt to the growing 3’ end RNA strand peels away & DNA helix reforms reads DNA 3 5
Transcription: Elongation Animations: http: //www- class. unl. edu/bioche m/gp 2/m_biology/an imation/gene_ a 2. html http: //vcell. ndsu. edu /animations/transcri ption/movieflash. htm

What would be the complementary RNA strand for the following DNA sequence? DNA 5’-GCGTATG-3’

Transcription: termination � Eukaryotes �RNA Polymerase transcribes a “signal” (AAUAAA) in terminator region of gene �END
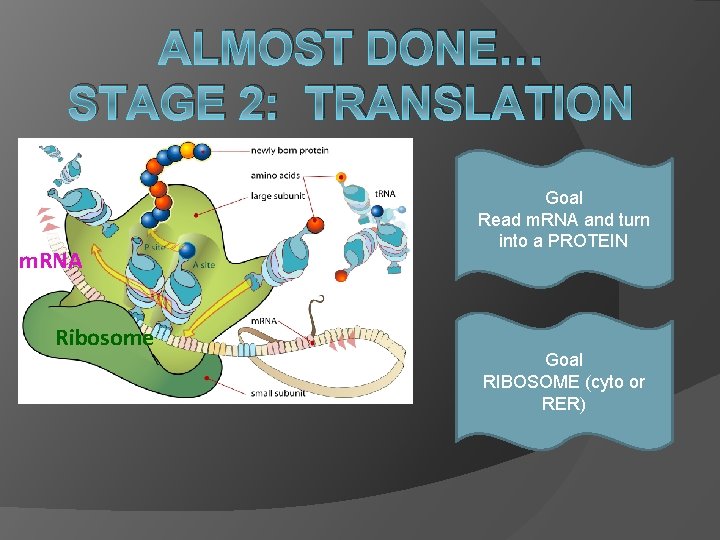
ALMOST DONE… STAGE 2: TRANSLATION m. RNA Ribosome Goal Read m. RNA and turn into a PROTEIN Goal RIBOSOME (cyto or RER)
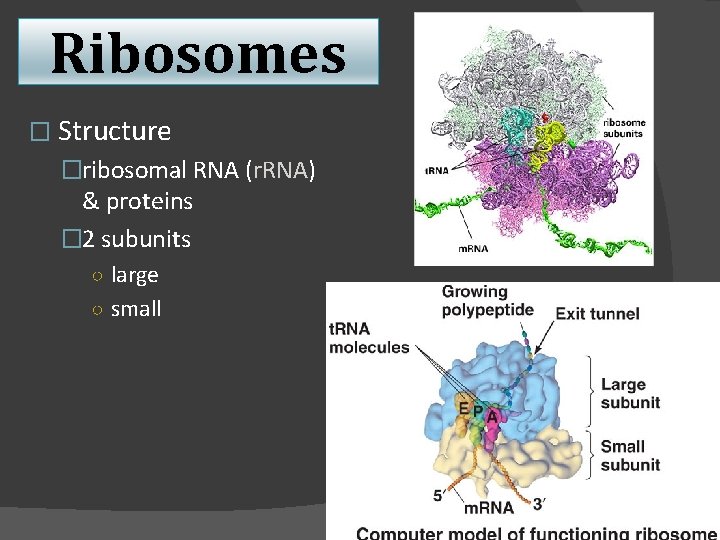
Ribosomes � Structure �ribosomal RNA (r. RNA) & proteins � 2 subunits ○ large ○ small
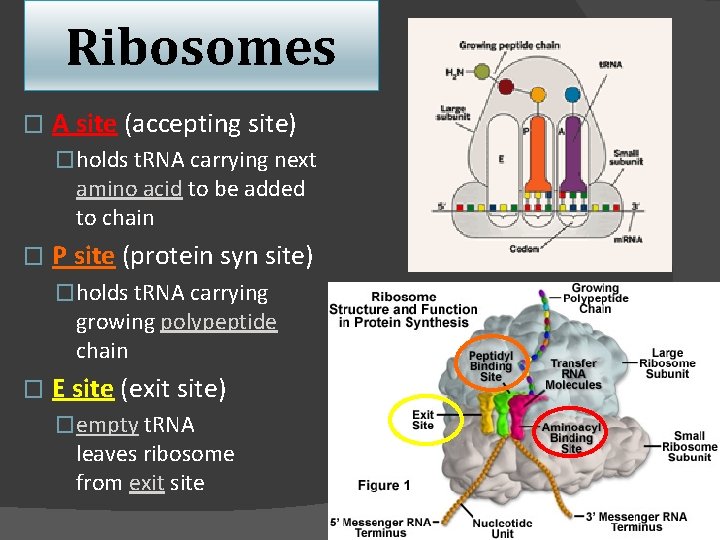
Ribosomes � A site (accepting site) �holds t. RNA carrying next amino acid to be added to chain � P site (protein syn site) �holds t. RNA carrying growing polypeptide chain � E site (exit site) �empty t. RNA leaves ribosome from exit site

Codon Chart m. RNA is read in sets of 3 bases…CODON codes for Amino Acid l l Start codon • • AUG methionine • UGA, UAG Stop codons
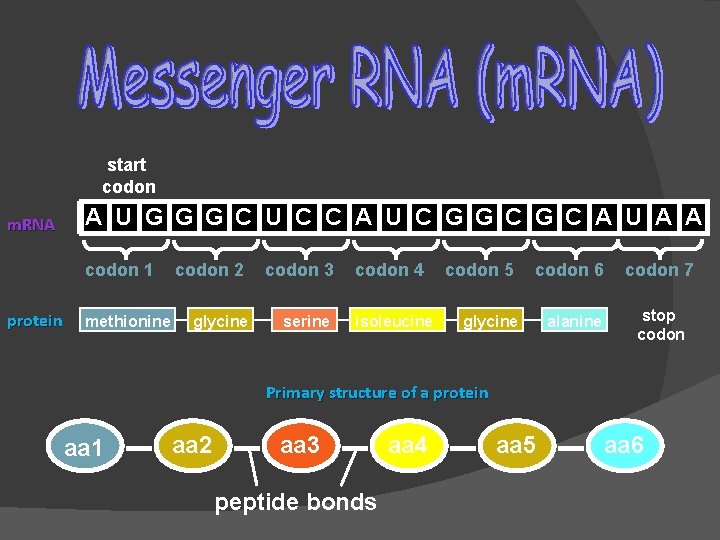
start codon m. RNA A U G G G C U C C A U C G G C A U A A codon 1 protein methionine codon 2 codon 3 glycine serine codon 4 isoleucine codon 5 codon 6 glycine alanine codon 7 stop codon Primary structure of a protein aa 1 aa 2 aa 3 peptide bonds aa 4 aa 5 aa 6

Yikes…here come the details!!
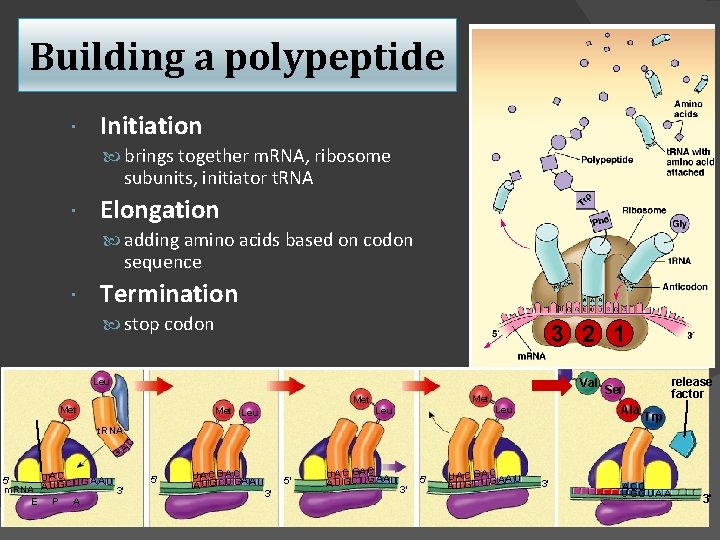
Building a polypeptide Initiation brings together m. RNA, ribosome subunits, initiator t. RNA Elongation adding amino acids based on codon sequence Termination stop codon 3 2 1 Val Leu Met Met Leu Ala Leu release factor Ser Trp A C t. RNA G U AC 5' CUGAA U m. RNA A U G 3' E P A 5' U A C G A C AA U G CU G 5' 3' U A C GA C A U G C UG AAU 3' 5' U A C G A C AA U AU G C U G 3' A CC U GG U A A 3'
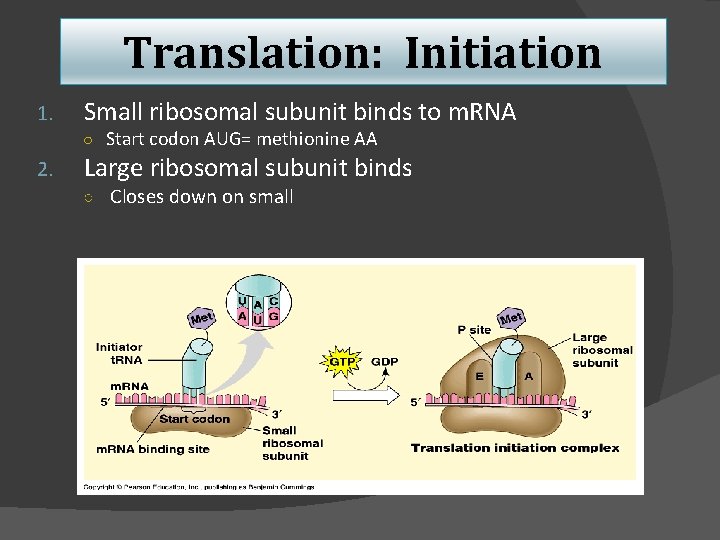
Translation: Initiation 1. Small ribosomal subunit binds to m. RNA ○ Start codon AUG= methionine AA 2. Large ribosomal subunit binds ○ Closes down on small
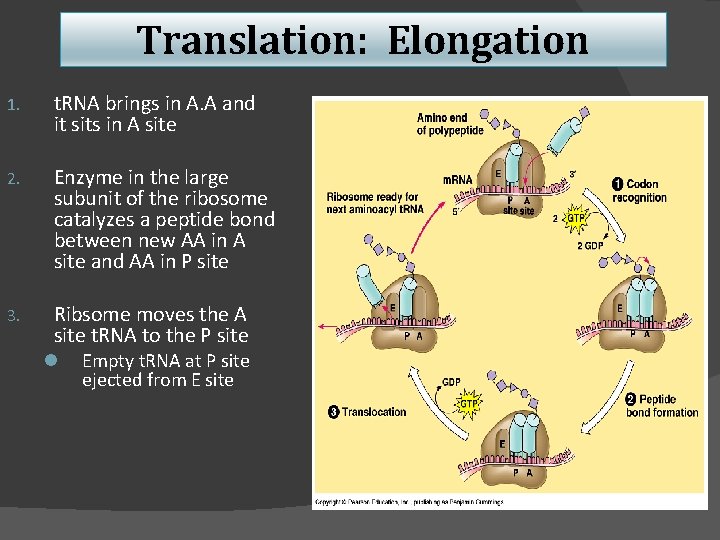
Translation: Elongation 1. t. RNA brings in A. A and it sits in A site 2. Enzyme in the large subunit of the ribosome catalyzes a peptide bond between new AA in A site and AA in P site 3. Ribsome moves the A site t. RNA to the P site l Empty t. RNA at P site ejected from E site
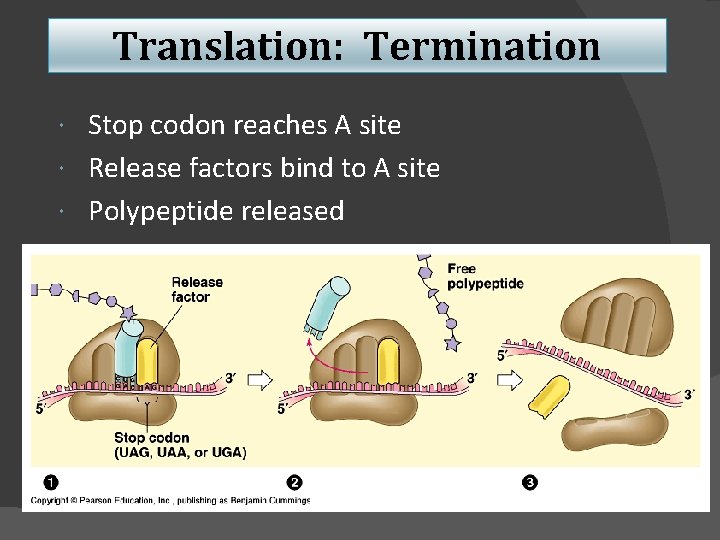
Translation: Termination Stop codon reaches A site Release factors bind to A site Polypeptide released
Translation Animations http: //vcell. ndsu. edu /animations/translati on/movie-flash. htm
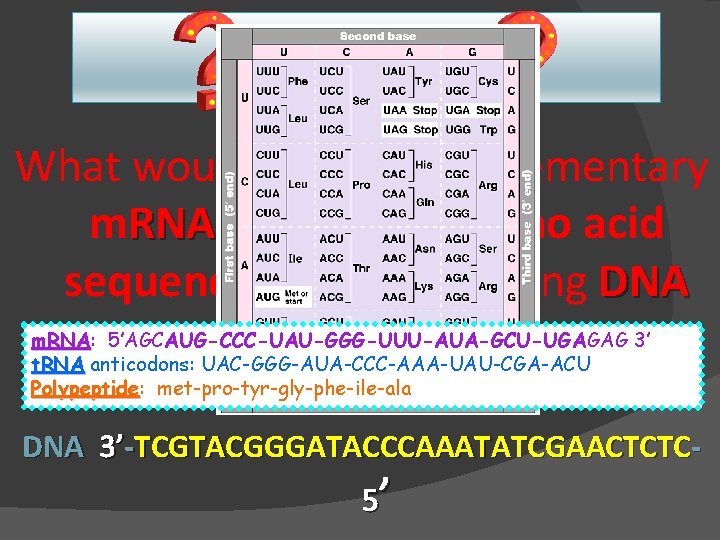
Question What would be the complementary m. RNA strand amino acid sequence for the following DNA m. RNA: 3’ m. RNA 5’AGCAUG-CCC-UAU-GGG-UUU-AUA-GCU-UGAGAG sequence? t. RNA anticodons: UAC-GGG-AUA-CCC-AAA-UAU-CGA-ACU Polypeptide: Polypeptide met-pro-tyr-gly-phe-ile-ala DNA 3’-TCGTACGGGATACCCAAATATCGAACTCTC 5’
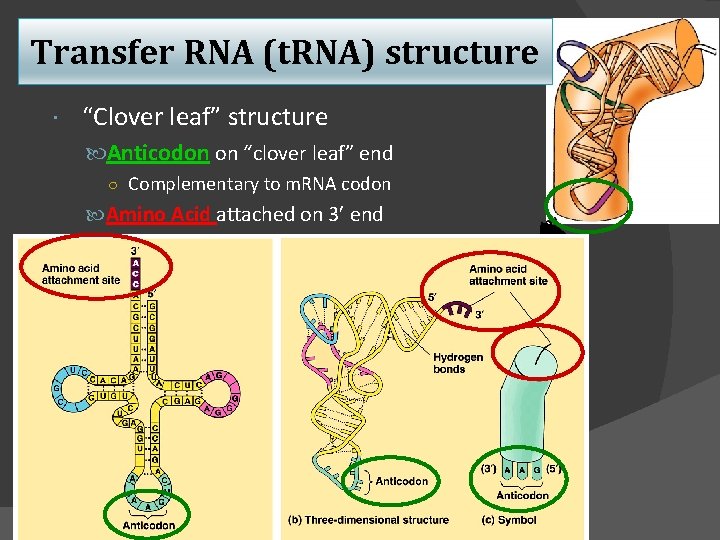
Transfer RNA (t. RNA) structure “Clover leaf” structure Anticodon on “clover leaf” end ○ Complementary to m. RNA codon Amino Acid attached on 3 end

Practice all of Protein Synthesis
Compare the 3 types of RNA

Prokaryote vs Eukaryote
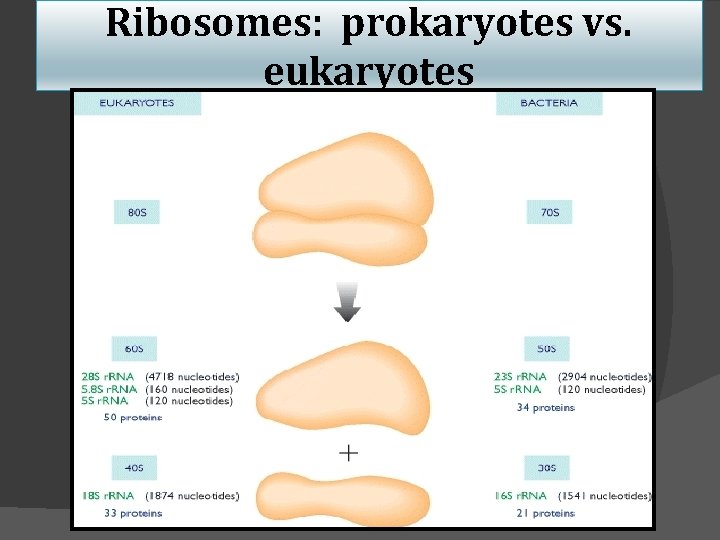
Ribosomes: prokaryotes vs. eukaryotes

Prokaryote Transcription initiation: RNA Polymerase recognizes promoter and binds…no transcription factors needed
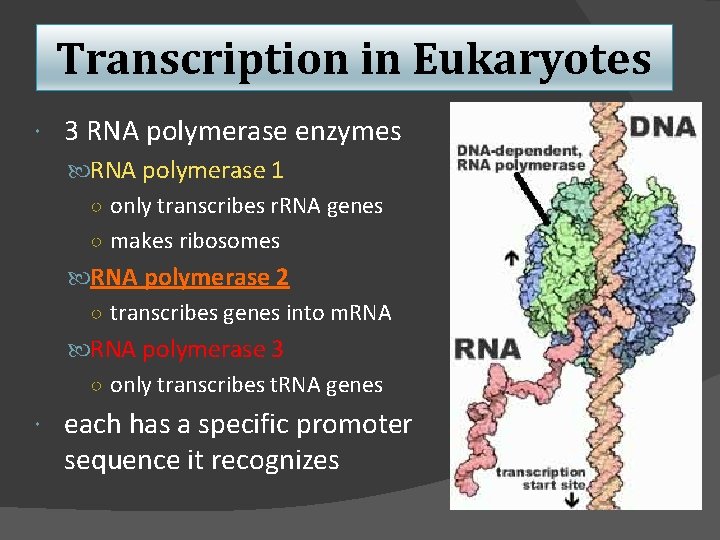
Transcription in Eukaryotes 3 RNA polymerase enzymes RNA polymerase 1 ○ only transcribes r. RNA genes ○ makes ribosomes RNA polymerase 2 ○ transcribes genes into m. RNA polymerase 3 ○ only transcribes t. RNA genes each has a specific promoter sequence it recognizes
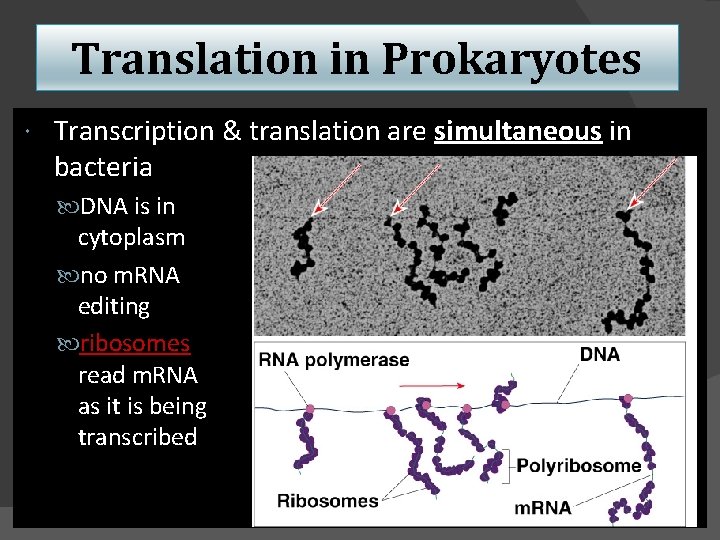
Translation in Prokaryotes Transcription & translation are simultaneous in bacteria DNA is in cytoplasm no m. RNA editing ribosomes read m. RNA as it is being transcribed

WHAT HAPPENS TO THE PROTEIN WHEN THE DNA IS WRONG? ?
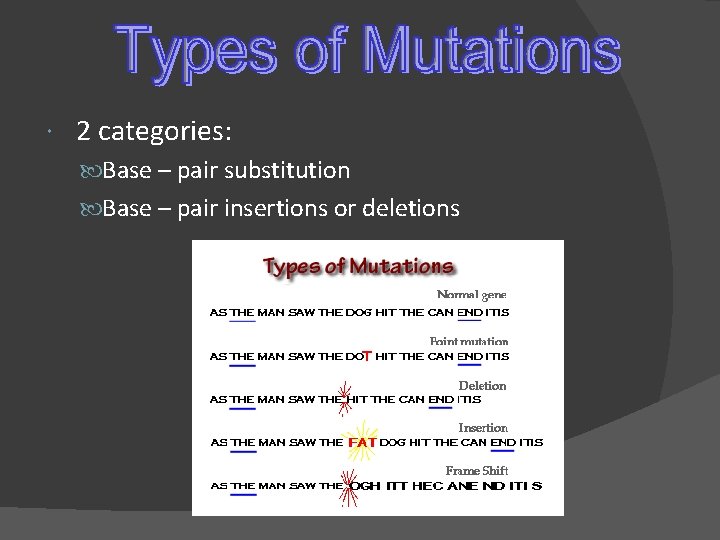
2 categories: Base – pair substitution Base – pair insertions or deletions
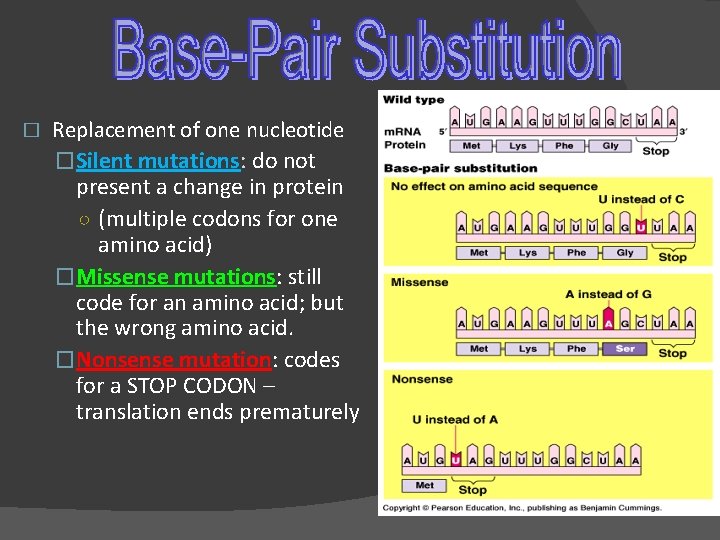
� Replacement of one nucleotide �Silent mutations: do not present a change in protein ○ (multiple codons for one amino acid) �Missense mutations: still code for an amino acid; but the wrong amino acid. �Nonsense mutation: codes for a STOP CODON – translation ends prematurely
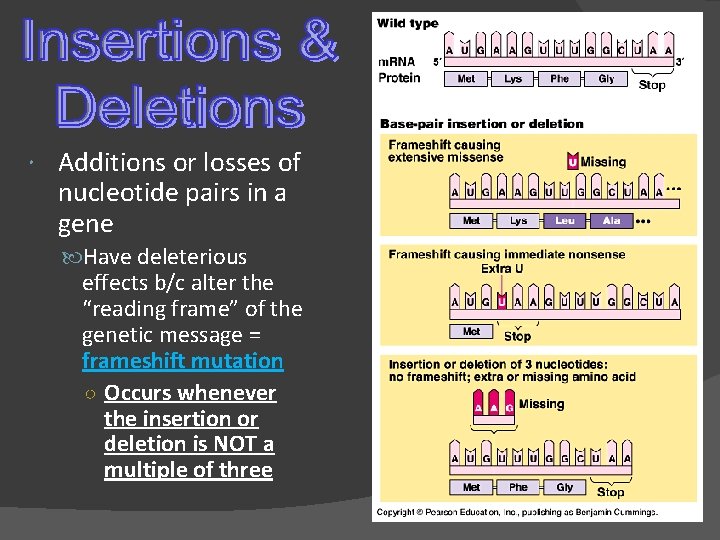
Additions or losses of nucleotide pairs in a gene Have deleterious effects b/c alter the “reading frame” of the genetic message = frameshift mutation ○ Occurs whenever the insertion or deletion is NOT a multiple of three
Regulating protein synthesis

Regulating at DNA level
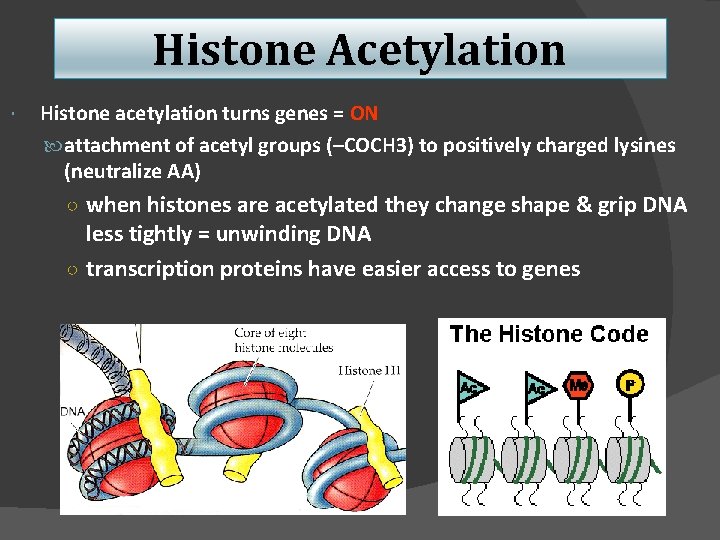
Histone Acetylation Histone acetylation turns genes = ON attachment of acetyl groups (–COCH 3) to positively charged lysines (neutralize AA) ○ when histones are acetylated they change shape & grip DNA less tightly = unwinding DNA ○ transcription proteins have easier access to genes
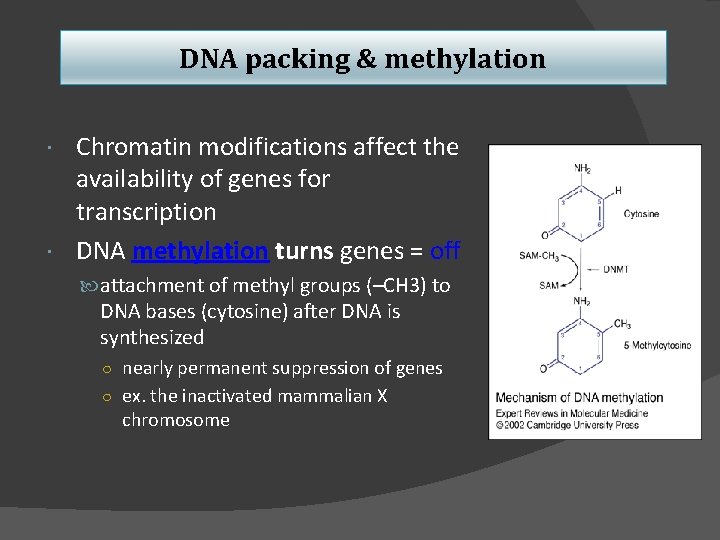
DNA packing & methylation Chromatin modifications affect the availability of genes for transcription DNA methylation turns genes = off attachment of methyl groups (–CH 3) to DNA bases (cytosine) after DNA is synthesized ○ nearly permanent suppression of genes ○ ex. the inactivated mammalian X chromosome
Regulating at transcription level
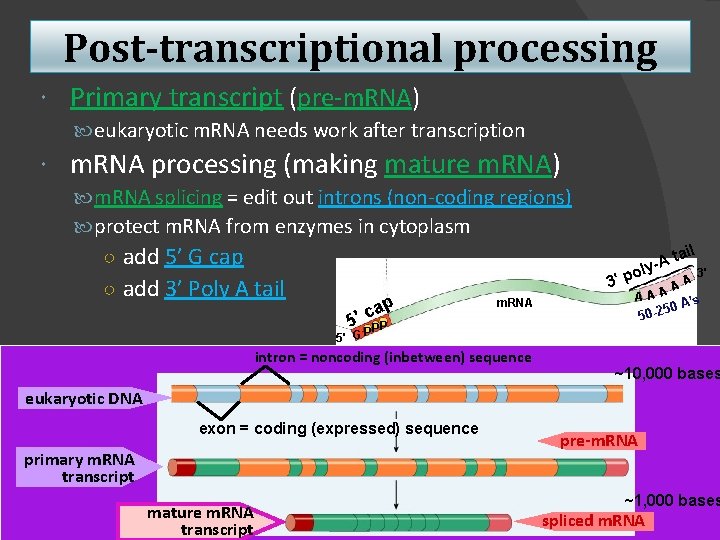
Post-transcriptional processing Primary transcript (pre-m. RNA) eukaryotic m. RNA needs work after transcription m. RNA processing (making mature m. RNA) m. RNA splicing = edit out introns (non-coding regions) protect m. RNA from enzymes in cytoplasm ○ add 5’ G cap ○ add 3’ Poly A tail il a t ly-A 3' o 3' p ap c ' 5 m. RNA AAA A s 0 A’ 5 50 -2 PP 5' G P intron = noncoding (inbetween) sequence A ~10, 000 bases eukaryotic DNA exon = coding (expressed) sequence primary m. RNA transcript mature m. RNA transcript pre-m. RNA ~1, 000 bases spliced m. RNA
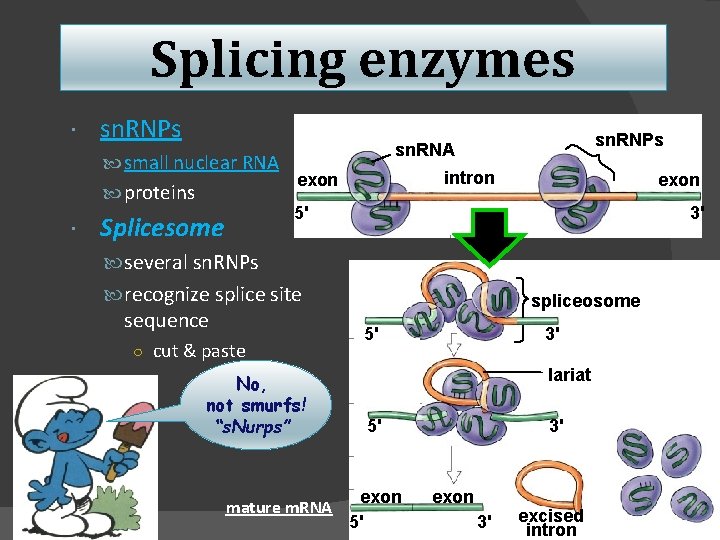
Splicing enzymes sn. RNPs small nuclear RNA proteins sn. RNPs sn. RNA intron exon 5' Splicesome 3' several sn. RNPs recognize splice site sequence ○ cut & paste No, not smurfs! “s. Nurps” mature m. RNA spliceosome 5' 3' lariat 5' exon 5' 3' exon 3' excised intron
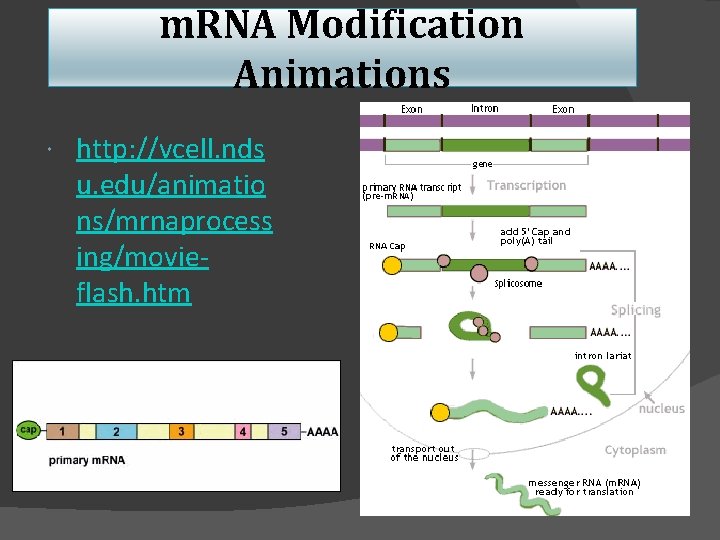
m. RNA Modification Animations http: //vcell. nds u. edu/animatio ns/mrnaprocess ing/movieflash. htm

Regulating at translation level
- Slides: 46