Protein Surface Analysis for Functional Analysis and Prediction






- Slides: 28

Protein Surface Analysis for Functional Analysis and Prediction T. Andrew Binkowski and Andrzej Joachimiak 2009 NIGMS Workshop: Enabling Technologies for Structural Biology March 4 -6, 2009

Outline How Can Surface Analysis Aid Your Structural Genomics Effort? § Protein Surfaces § Comparing Surfaces of Proteins § Surface Analysis in the Structural Genomics Pipeline § The Global Protein Surface Survey

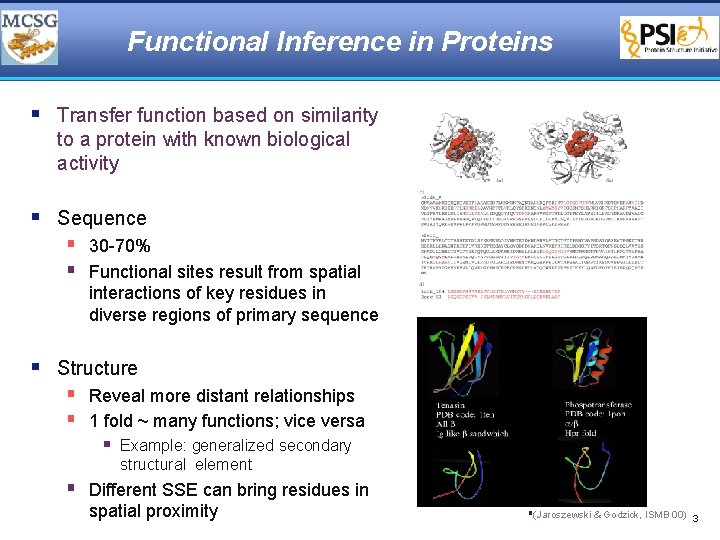
Functional Inference in Proteins § Transfer function based on similarity to a protein with known biological activity § Sequence § 30 -70% § Functional sites result from spatial interactions of key residues in diverse regions of primary sequence § Structure § Reveal more distant relationships § 1 fold ~ many functions; vice versa § Example: generalized secondary structural element § Different SSE can bring residues in spatial proximity §(Jaroszewski & Godzick, ISMB 00) 3

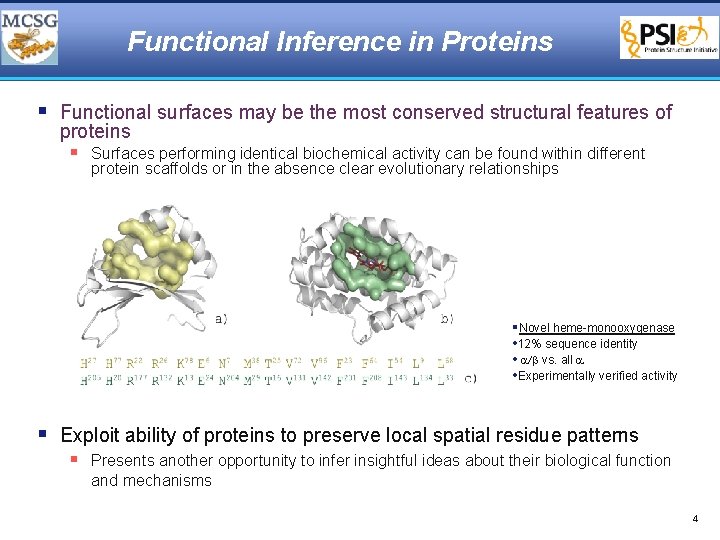
Functional Inference in Proteins § Functional surfaces may be the most conserved structural features of proteins § Surfaces performing identical biochemical activity can be found within different protein scaffolds or in the absence clear evolutionary relationships §Novel heme-monooxygenase • 12% sequence identity • a/b vs. all a • Experimentally verified activity § Exploit ability of proteins to preserve local spatial residue patterns § Presents another opportunity to infer insightful ideas about their biological function and mechanisms 4

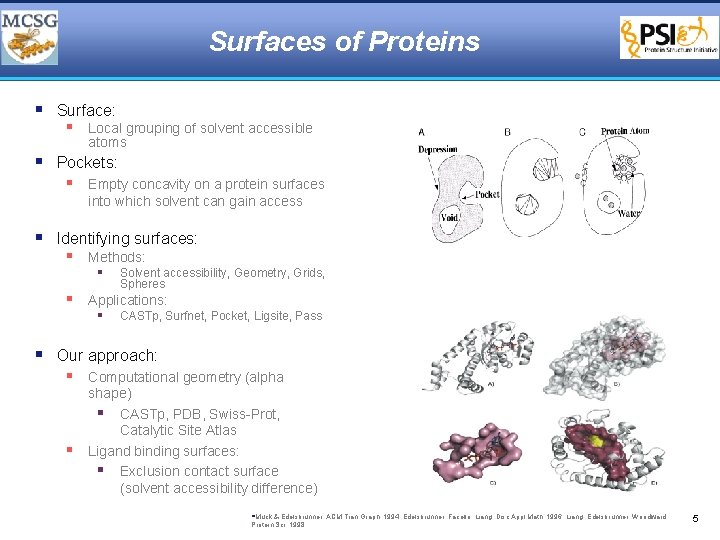
Surfaces of Proteins § Surface: § Local grouping of solvent accessible atoms § Pockets: § Empty concavity on a protein surfaces into which solvent can gain access § Identifying surfaces: § Methods: § Solvent accessibility, Geometry, Grids, Spheres § Applications: § CASTp, Surfnet, Pocket, Ligsite, Pass § Our approach: § Computational geometry (alpha § shape) § CASTp, PDB, Swiss-Prot, Catalytic Site Atlas Ligand binding surfaces: § Exclusion contact surface (solvent accessibility difference) §Muck & Edelsbrunner, ACM Tran Graph, 1994; Edelsbrunner, Facello, Liang, Disc Appl Math, 1996; Liang, Edelsbrunner, Woodward, Protein Sci, 1998 5

Global Protein Surface Survey http: //gpss. mcsg. anl. gov 6

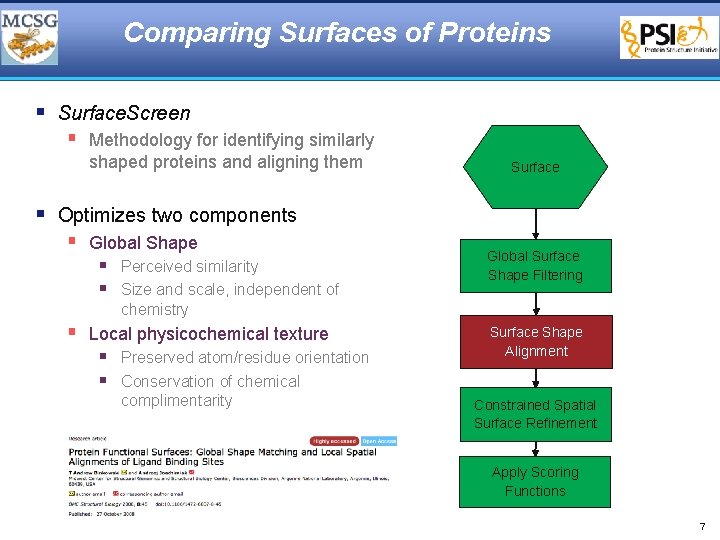
Comparing Surfaces of Proteins § Surface. Screen § Methodology for identifying similarly shaped proteins and aligning them Surface § Optimizes two components § Global Shape § Perceived similarity § Size and scale, independent of Global Surface Shape Filtering chemistry § Local physicochemical texture § Preserved atom/residue orientation § Conservation of chemical complimentarity Surface Shape Alignment Constrained Spatial Surface Refinement Apply Scoring Functions 7

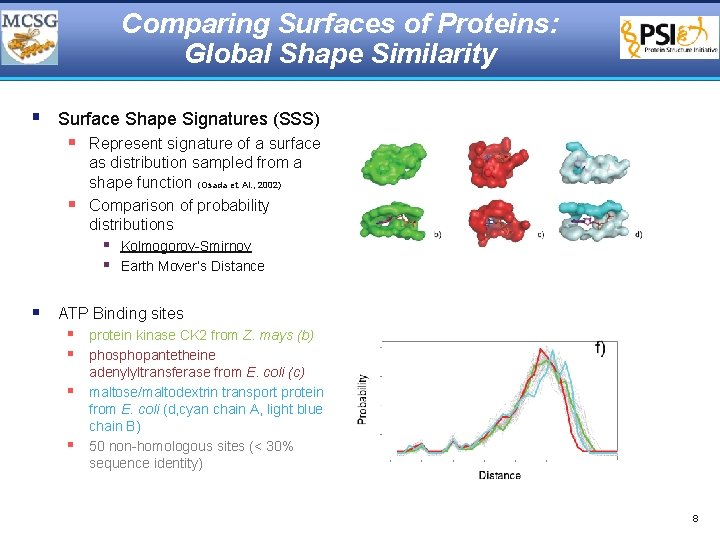
Comparing Surfaces of Proteins: Global Shape Similarity § Surface Shape Signatures (SSS) § Represent signature of a surface § as distribution sampled from a shape function (Osada et. Al. , 2002) Comparison of probability distributions § Kolmogorov-Smirnov § Earth Mover’s Distance § ATP Binding sites § protein kinase CK 2 from Z. mays (b) § phosphopantetheine § § adenylyltransferase from E. coli (c) maltose/maltodextrin transport protein from E. coli (d, cyan chain A, light blue chain B) 50 non-homologous sites (< 30% sequence identity) 8

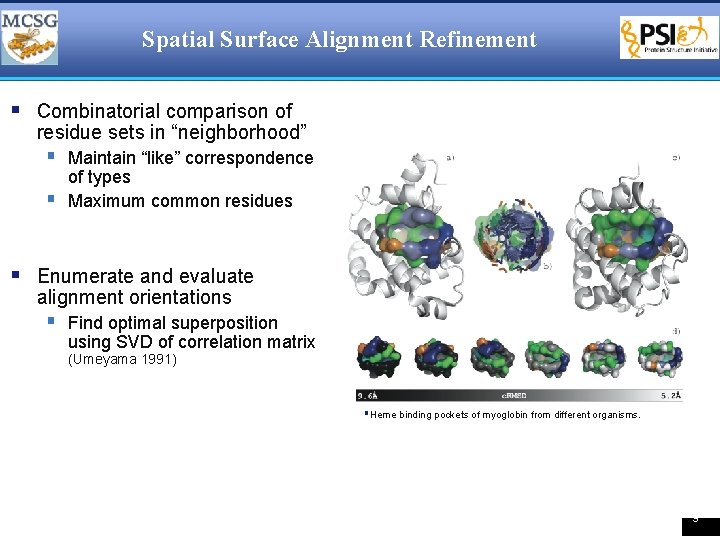
Spatial Surface Alignment Refinement § Combinatorial comparison of residue sets in “neighborhood” § Maintain “like” correspondence § of types Maximum common residues § Enumerate and evaluate alignment orientations § Find optimal superposition using SVD of correlation matrix (Umeyama 1991) §Heme binding pockets of myoglobin from different organisms. 9

Evaluating Surface Alignments § RMSD Distance: § Estimate the probability of obtaining a specific RMSD for nres § Compute random surface alignments (108) and build lookup tables § RMSD variants: § § c. RMSD (coordinate) o. RMSD (orientation) § Surface Volume Overlap: § Interpretation of SVOT is not straightforward § Need global and local 10

Benchmarking Surface Alignments 11

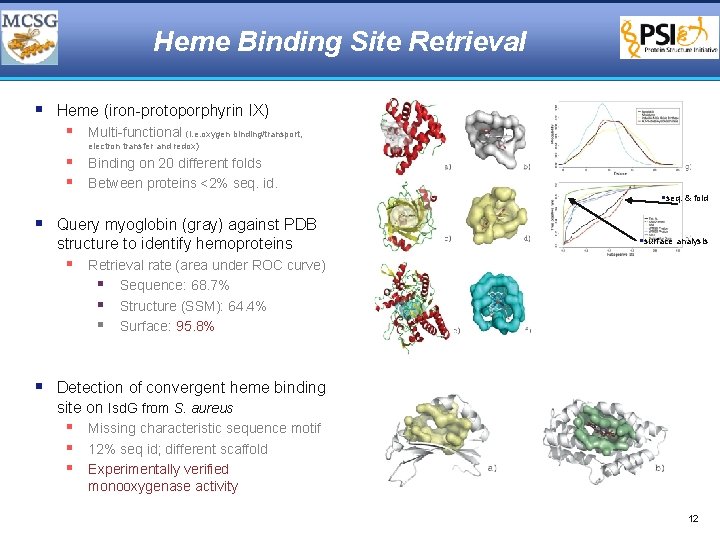
Heme Binding Site Retrieval § Heme (iron-protoporphyrin IX) § Multi-functional (i. e. oxygen binding/transport, electron transfer and redox) § Binding on 20 different folds § Between proteins <2% seq. id. §seq. & fold § Query myoglobin (gray) against PDB structure to identify hemoproteins §surface analysis § Retrieval rate (area under ROC curve) § Sequence: 68. 7% § Structure (SSM): 64. 4% § Surface: 95. 8% § Detection of convergent heme binding site on Isd. G from S. aureus § Missing characteristic sequence motif § 12% seq id; different scaffold § Experimentally verified monooxygenase activity 12

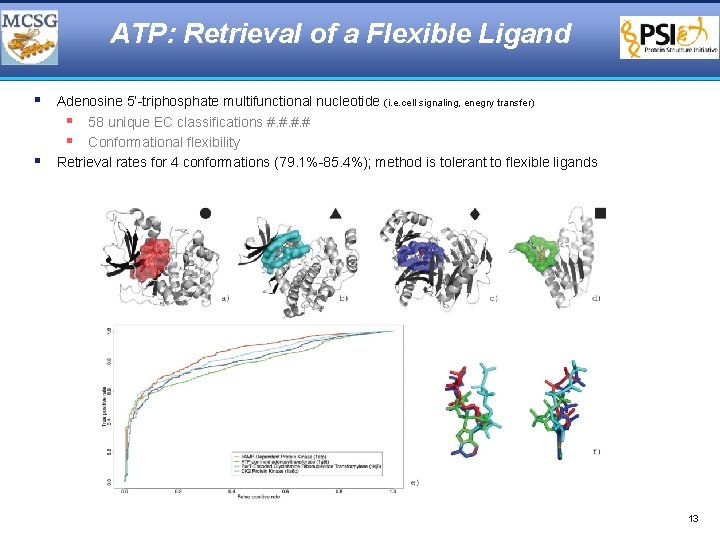
ATP: Retrieval of a Flexible Ligand § Adenosine 5’-triphosphate multifunctional nucleotide (i. e. cell signaling, enegry transfer) § 58 unique EC classifications #. # § Conformational flexibility § Retrieval rates for 4 conformations (79. 1%-85. 4%); method is tolerant to flexible ligands 13

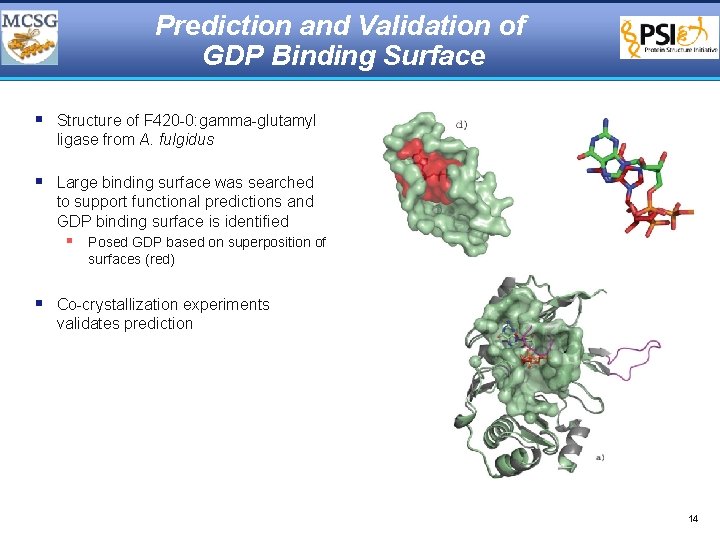
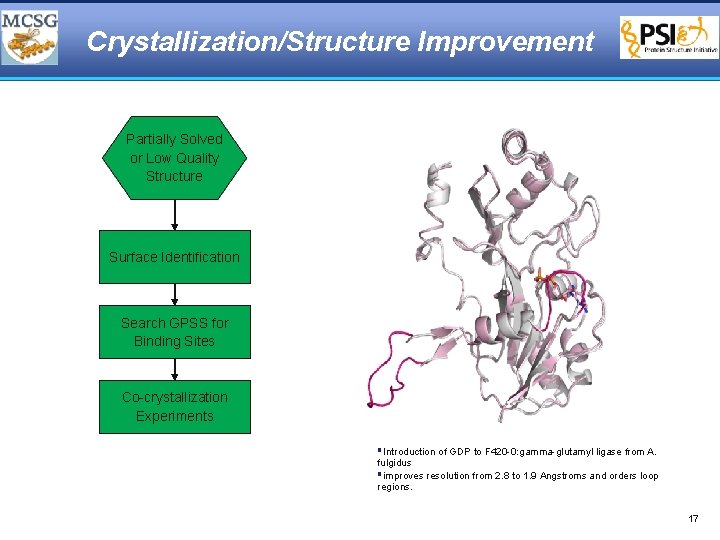
Prediction and Validation of GDP Binding Surface § Structure of F 420 -0: gamma-glutamyl ligase from A. fulgidus § Large binding surface was searched to support functional predictions and GDP binding surface is identified § Posed GDP based on superposition of surfaces (red) § Co-crystallization experiments validates prediction 14

Surface Analysis in the Structural Genomics Pipeline 15

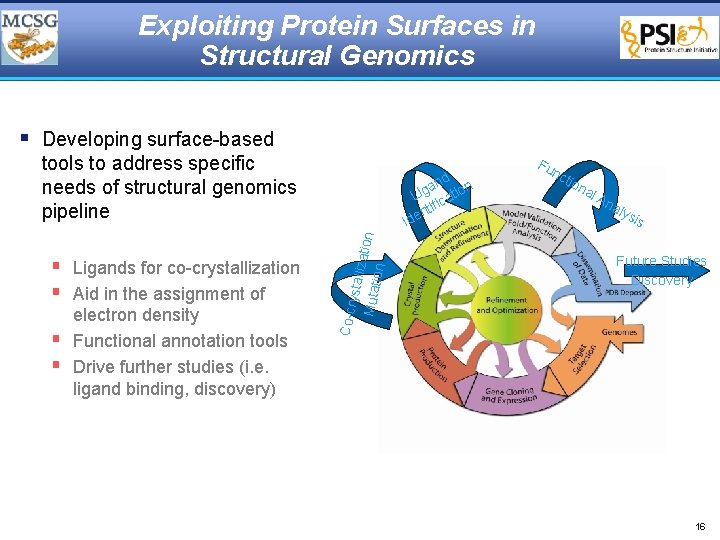
Exploiting Protein Surfaces in Structural Genomics § Developing surface-based tools to address specific needs of structural genomics pipeline § § ystall iz Mutat ation electron density Functional annotation tools Drive further studies (i. e. ligand binding, discovery) Fu nc tio na l. A na lys is Future Studies Discovery Co-cr § Ligands for co-crystallization § Aid in the assignment of nd n a g Li catio fi nti e Id 16

Crystallization/Structure Improvement Partially Solved or Low Quality Structure Surface Identification Search GPSS for Binding Sites Co-crystallization Experiments §Introduction of GDP to F 420 -0: gamma-glutamyl ligase from A. fulgidus §improves resolution from 2. 8 to 1. 9 Angstroms and orders loop regions. 17

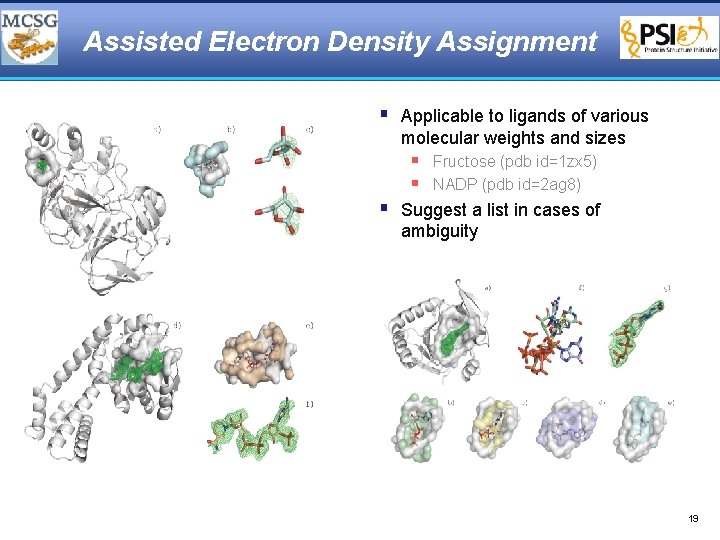
Assisted Electron Density Assignment § Unidentified ligand density § Construct surface surrounding density and search against ligand surface library § Does not require entire structure to be built 18

Assisted Electron Density Assignment § Applicable to ligands of various molecular weights and sizes § Fructose (pdb id=1 zx 5) § NADP (pdb id=2 ag 8) § Suggest a list in cases of ambiguity 19

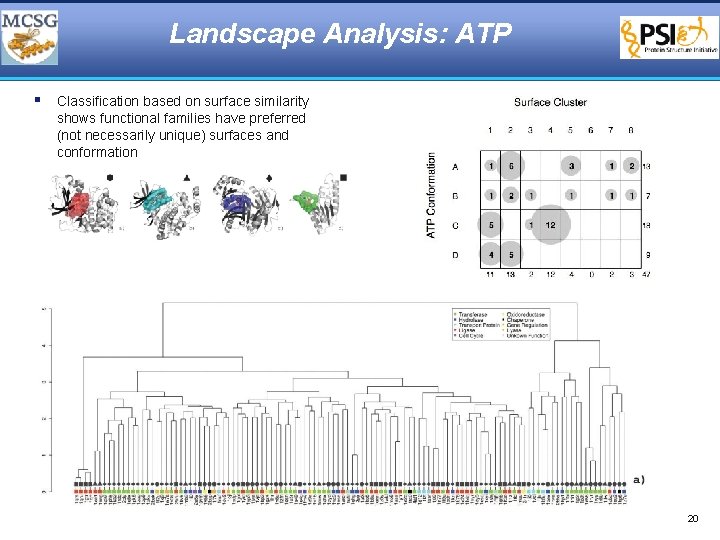
Landscape Analysis: ATP § Classification based on surface similarity shows functional families have preferred (not necessarily unique) surfaces and conformation 20

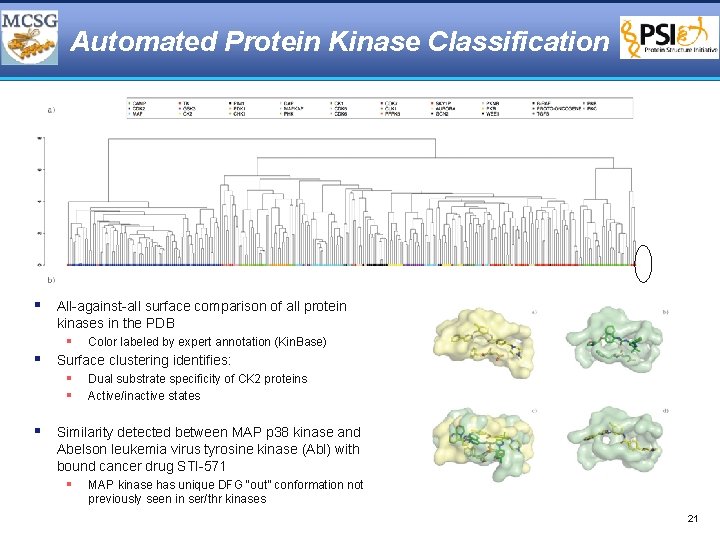
Automated Protein Kinase Classification § All-against-all surface comparison of all protein kinases in the PDB § Color labeled by expert annotation (Kin. Base) § Surface clustering identifies: § § Dual substrate specificity of CK 2 proteins Active/inactive states § Similarity detected between MAP p 38 kinase and Abelson leukemia virus tyrosine kinase (Abl) with bound cancer drug STI-571 § MAP kinase has unique DFG “out” conformation not previously seen in ser/thr kinases 21

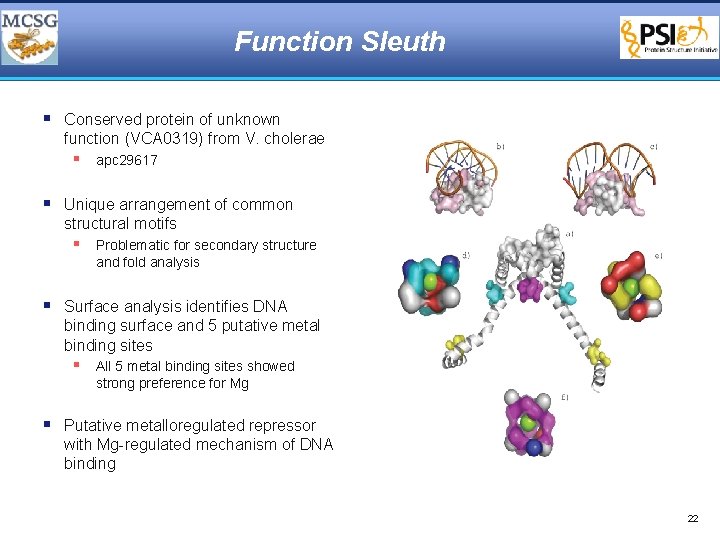
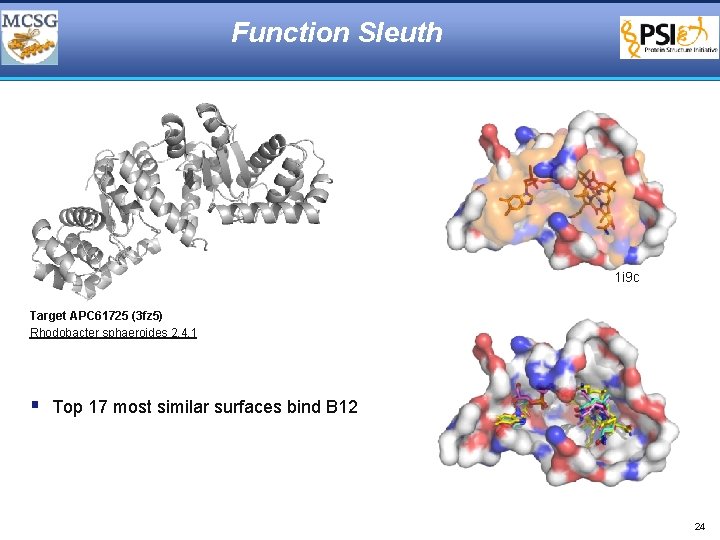
Function Sleuth § Conserved protein of unknown function (VCA 0319) from V. cholerae § apc 29617 § Unique arrangement of common structural motifs § Problematic for secondary structure and fold analysis § Surface analysis identifies DNA binding surface and 5 putative metal binding sites § All 5 metal binding sites showed strong preference for Mg § Putative metalloregulated repressor with Mg-regulated mechanism of DNA binding 22

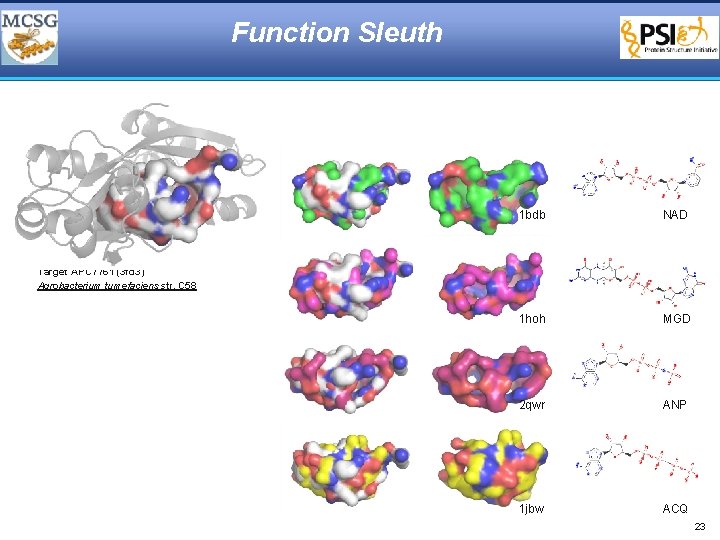
Function Sleuth 1 bdb NAD 1 hoh MGD 2 qwr ANP 1 jbw ACQ Target APC 7761 (3 fd 3) Agrobacterium tumefaciens str. C 58 23

Function Sleuth 1 i 9 c Target APC 61725 (3 fz 5) Rhodobacter sphaeroides 2. 4. 1 § Top 17 most similar surfaces bind B 12 24

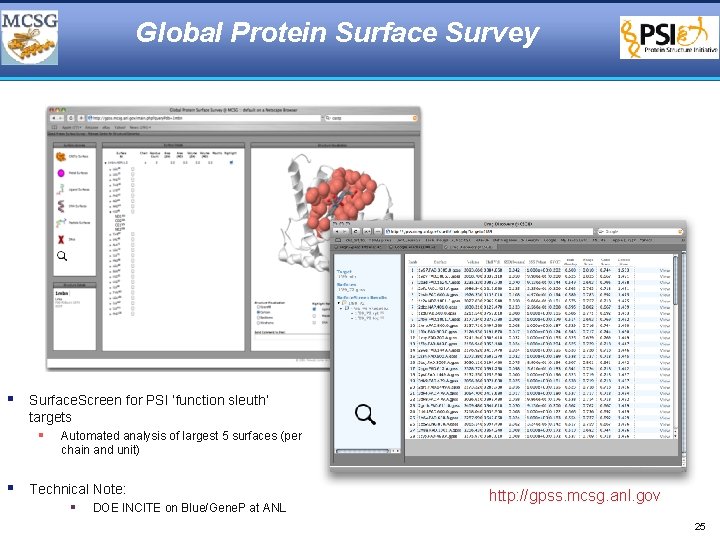
Global Protein Surface Survey § Surface. Screen for PSI ‘function sleuth’ targets § Automated analysis of largest 5 surfaces (per chain and unit) § Technical Note: § DOE INCITE on Blue/Gene. P at ANL http: //gpss. mcsg. anl. gov 25

Conclusion § Comparing surfaces of proteins can be a useful tool with many applications § Functional characterization § Assisted electron density assignment § Automated classification § Global Protein Surface Survey § http: //gpss. mcsg. anl. gov 26

Acknowledgements ANL/MCSG H. An, G. Babnigg, L. Bigelow, A. Binkowski, C-s. Chang, S. Clancy, G. Cobb, M. Cuff, M. Donnelly, C. Giometti, W. Eschenfeldt, Y. Fan, C. Hatzos, R. Hendricks G. Joachimiak, H. Li, L. Keigher, Y-c. Kim, N. Maltseva, E. Marland, S. Moy, R. Mulligan, B. Nocek, J. Osipiuk, M. Schiffer, ANL/MCSG A. Sather, G. Shackelford, L. Stols, K. Tan, C. Tesar, R-y. Wu, L. Volkart, R-g. Zhang, M. Zhou, ANL/SBC N. Duke, S. Ginell, F. Rotella Univ. of Virginia W. Minor, M. Chruszcz, M. Cyborowski, M. Grabowski, P. Lasota, P. Miles, M. Zimmerman, H. Zheng Univ. College London @ EBI, J. Thornton, C. Orengo, M. Bashton, R. Laskowski, D. Lee, R. Marsden, D. Mc. Kenzie, A. Todd, J. Watson Northwestern Univ. W. Anderson, O. Kiryukhina D. Miller, G. Minasov, L. Shuvalova, X. Yang, Y. Tang G. Montelione, Ruthgers Univ. NESGC T. Terwilliger, Los Alamos, ITCSG Z. Derewenda, Univ. of Virginia, ITCSG Z. Dauter, NCI J. Liang, Univ. of Illinois D. Sherman, U. Michigan Washington Univ. D. Fremont, T. Brett, C. Nelson, Univ. of Texas SWMC Z. Otwinowski, D. Borek, A. Kudlicki, A. Q. Mei, M. Rowicka Funding: NIH and DOE Univ. of Toronto A. Edwards, C. Arrowsmith, A. Savchenko, E. Evdokimova, J. Guthrie, A. Khachatryan, M. Kudrytska, T. Skarina, X. (Linda) Xu Univ. of Chicago O. Schneewind, D. Missiakas, P. Gornicki, S. Koide, ITCSG W-j. Tang, B. Roux, J. L. Robertson M. R. Rosner, T. Kossiakoff, ITCSG V. Tereshko, 27

Thank you