Protein Structures Data Representation 1 Primary Structure Structure





- Slides: 22

Protein Structures: Data Representation 1) Primary Structure: Structure character string. 2) Secondary Structure: Structure Identifying sub-structures in a large protein based on sequence. 3) Tertiary Structure: Structure 4) Quaternary Structure: Structure 3 -Dimensional Representation Protein Database Bank (PDB) This is a complicated file format structure that support numerous programs, and contains information regarding the primary structure (sequence), 3 -D structures (x, y, z coordinates), size and linking of specific atoms in structures, etc.

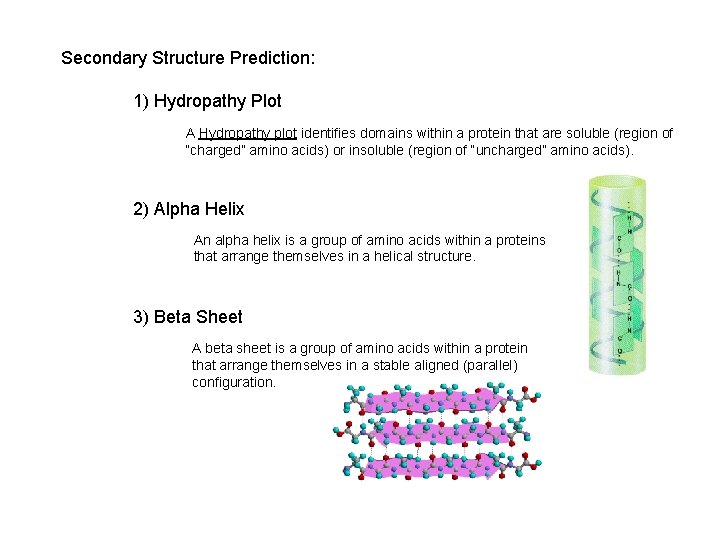
Secondary Structure Prediction: 1) Hydropathy Plot A Hydropathy plot identifies domains within a protein that are soluble (region of “charged” amino acids) or insoluble (region of “uncharged” amino acids). 2) Alpha Helix An alpha helix is a group of amino acids within a proteins that arrange themselves in a helical structure. 3) Beta Sheet A beta sheet is a group of amino acids within a protein that arrange themselves in a stable aligned (parallel) configuration.



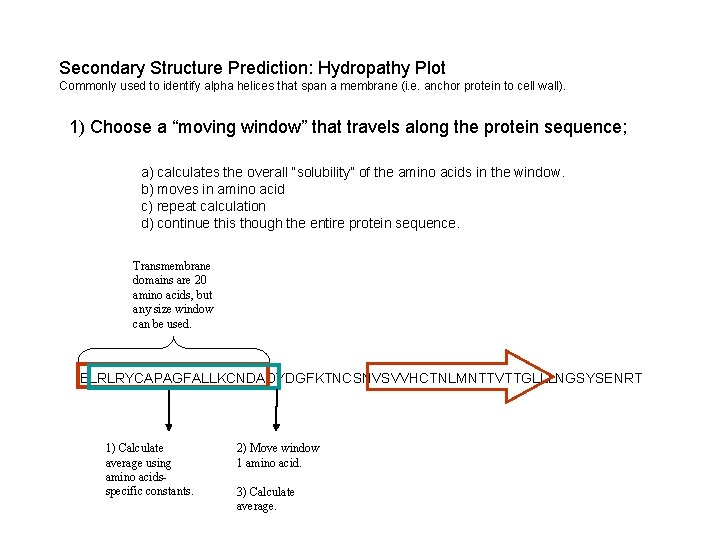
Secondary Structure Prediction: Hydropathy Plot Commonly used to identify alpha helices that span a membrane (i. e. anchor protein to cell wall). 1) Choose a “moving window” that travels along the protein sequence; a) calculates the overall “solubility” of the amino acids in the window. b) moves in amino acid c) repeat calculation d) continue this though the entire protein sequence. Transmembrane domains are 20 amino acids, but any size window can be used. ELRLRYCAPAGFALLKCNDADYDGFKTNCSNVSVVHCTNLMNTTVTTGLLLNGSYSENRT 1) Calculate average using amino acidsspecific constants. 2) Move window 1 amino acid. 3) Calculate average.

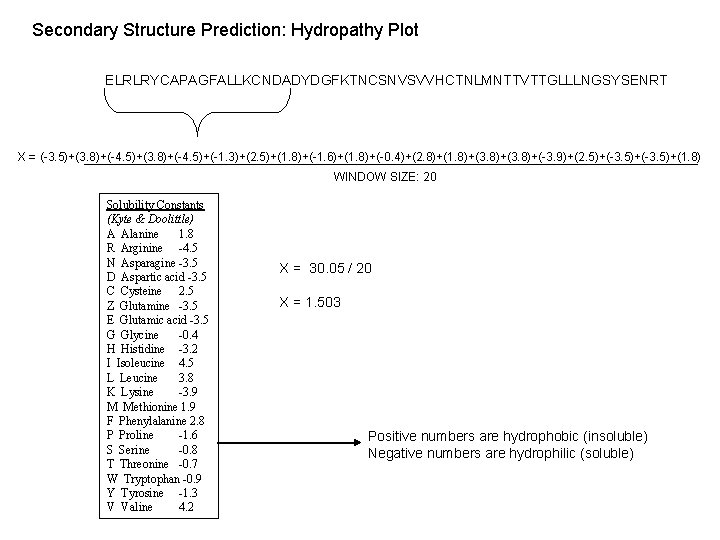
Secondary Structure Prediction: Hydropathy Plot ELRLRYCAPAGFALLKCNDADYDGFKTNCSNVSVVHCTNLMNTTVTTGLLLNGSYSENRT X = (-3. 5)+(3. 8)+(-4. 5)+(-1. 3)+(2. 5)+(1. 8)+(-1. 6)+(1. 8)+(-0. 4)+(2. 8)+(1. 8)+(3. 8)+(-3. 9)+(2. 5)+(-3. 5)+(1. 8) WINDOW SIZE: 20 Solubility Constants (Kyte & Doolittle) A Alanine 1. 8 R Arginine -4. 5 N Asparagine -3. 5 D Aspartic acid -3. 5 C Cysteine 2. 5 Z Glutamine -3. 5 E Glutamic acid -3. 5 G Glycine -0. 4 H Histidine -3. 2 I Isoleucine 4. 5 L Leucine 3. 8 K Lysine -3. 9 M Methionine 1. 9 F Phenylalanine 2. 8 P Proline -1. 6 S Serine -0. 8 T Threonine -0. 7 W Tryptophan -0. 9 Y Tyrosine -1. 3 V Valine 4. 2 X = 30. 05 / 20 X = 1. 503 Positive numbers are hydrophobic (insoluble) Negative numbers are hydrophilic (soluble)

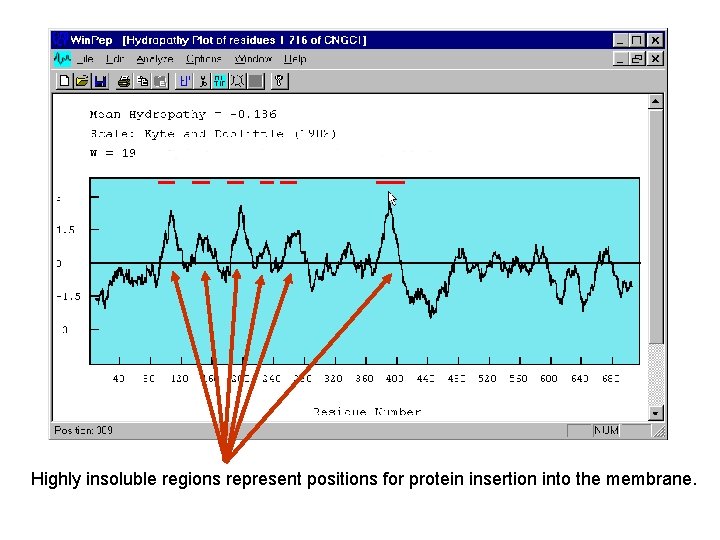
Highly insoluble regions represent positions for protein insertion into the membrane.

Protein Folding: Computationally Modeling Biochemistry

OBJECTIVE: Utilize the sequence information, along with temperaturedependent biomolecular interaction constants, to computationally “predict” a protein’s tertiary structure. CHALLENGES: A) It is NOT known how proteins fold in nature. B) More detailed or mathematically-intensive methods can’t be completed in a reasonable time (given current computer capabilities). C) There are essentially no experimental methods to verify or validate that a predicted protein is “correct” – or “how correct”.

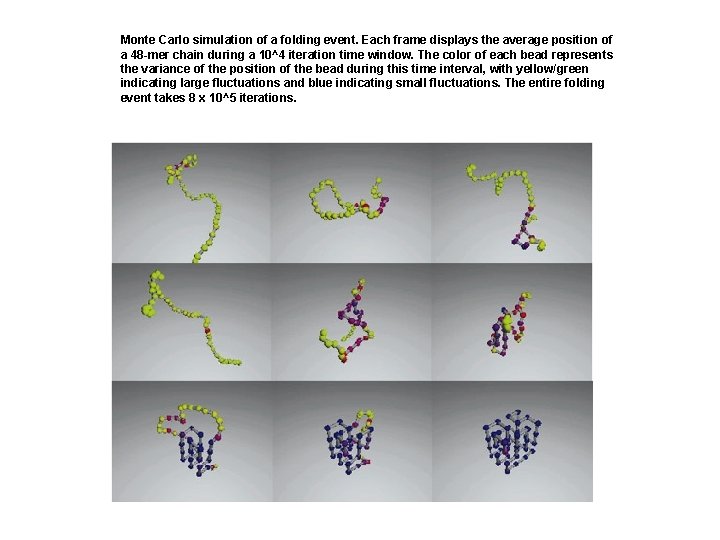
Monte Carlo simulation of a folding event. Each frame displays the average position of a 48 -mer chain during a 10^4 iteration time window. The color of each bead represents the variance of the position of the bead during this time interval, with yellow/green indicating large fluctuations and blue indicating small fluctuations. The entire folding event takes 8 x 10^5 iterations.

Evolution of Protein Folding Methods: 1) Lattice Methods: 3 D lattice of residue or atomic positions. 2) Off-Lattice Methods: Not reliant on predetermined 3 D positions. Can include solvent effects. 3) All Atoms Methods/Modeling: EXTREMELY computationally intensive. Tactics A) Initially calculate secondary structures minimums (fold sheets and helices), then calculate minima for remaining sequence. B) Emulate Protein synthesis process, starting from amino-terminus. C) Utilize existing NMR and X-ray crystal structures that match sequence under investigation.

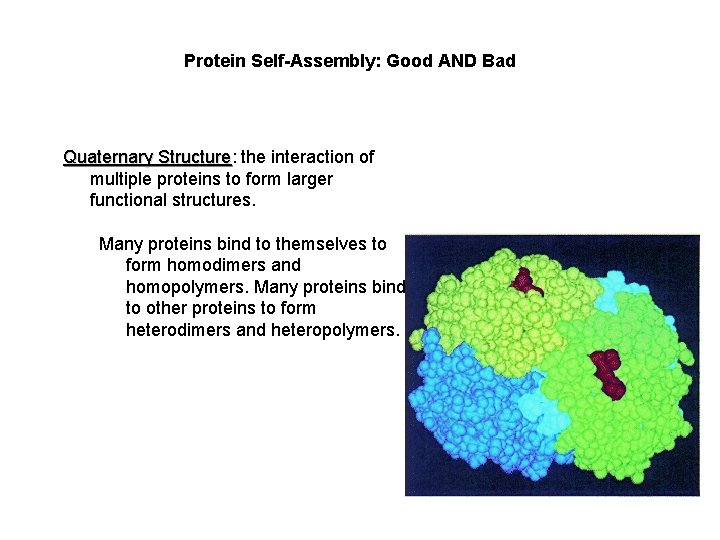
Protein Self-Assembly: Good AND Bad Quaternary Structure: Structure the interaction of multiple proteins to form larger functional structures. Many proteins bind to themselves to form homodimers and homopolymers. Many proteins bind to other proteins to form heterodimers and heteropolymers.

Many diseases involve self-aggregating proteins (especially neurodegenerative diseases). Mad Cow Disease (Prion Proteins) Alzheimer’s Disease (beta-Amyloid Peptide) Huntington’s Disease Why neuro-diseases? 1) Because the blood flow (nutrients) to the brain is highly regulated, and proteins that aggregate tend to collect – and are NEUROTOXIC. Note that these proteins ALSO aggregate in peripheral tissues, but are “cleared” and do not appear to be sufficiently toxic. 2) Brain cells (neurons) do NOT regenerate in a manner equivalent to peripheral tissues (particularly in older people). 3) Loss of neuronal cells leads to altered cognitive capabilities, which is not the case in peripheral tissues (e. g. slight muscle atrophy).

Neurodegenerative Protein Diseases = Beta Sheet Structures!!! Beta-sheet structures are sometimes called “amyloid” structures. Hence the term: Amyloidopathy NOTE: The molecular forces that assemble betasheet structures ALSO cause them to self-assemble!

2 key concepts regarding age-related diseases…. 1) Increased human health & longevity “invents diseases”. Before the modern age, nature had rarely seen a 60 year old human. Imagine the age-related diseases of the future when the average human life span is >120 years. 2) Evolutionary pressures did not select for humans to live much longer than 35 -40 years. So inherited mutations that lead to age-related diseases were not “selected out” of the human population. This fact has NOT changed in modern times. Alzheimer’s Disease 40 -90 (sporadic at 60+, familial at 40+), increases with age Men more common under the age of 80 yrs Women more common over the age of 80 yrs (J Neurol Neurosurg Psychiatry 1999; 66; 177 in BMJ 1999 Feb 27; 318(7183); 614)

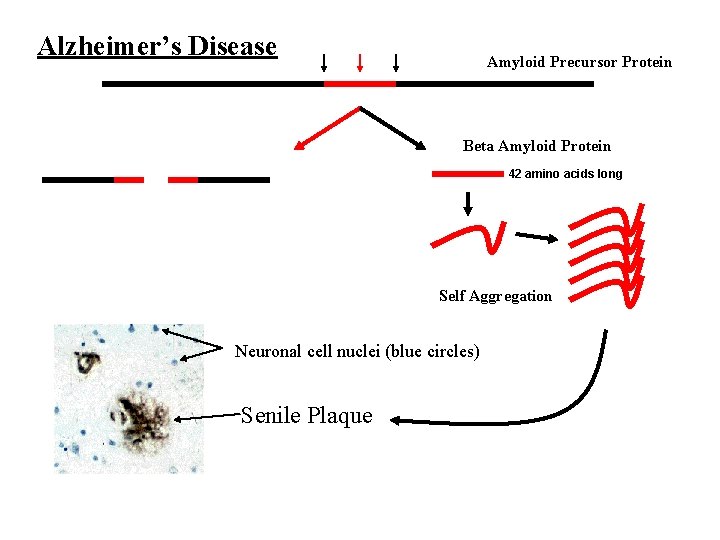
Alzheimer’s Disease Amyloid Precursor Protein Beta Amyloid Protein 42 amino acids long Self Aggregation Neuronal cell nuclei (blue circles) Senile Plaque

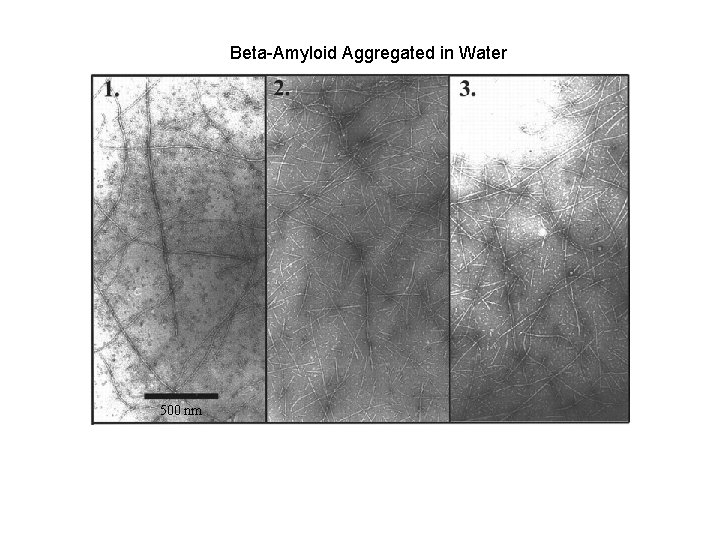
Beta-Amyloid Aggregated in Water 500 nm

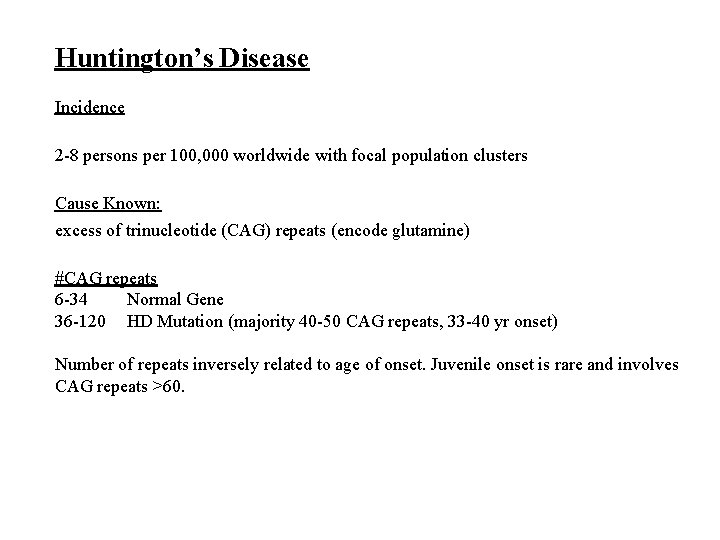
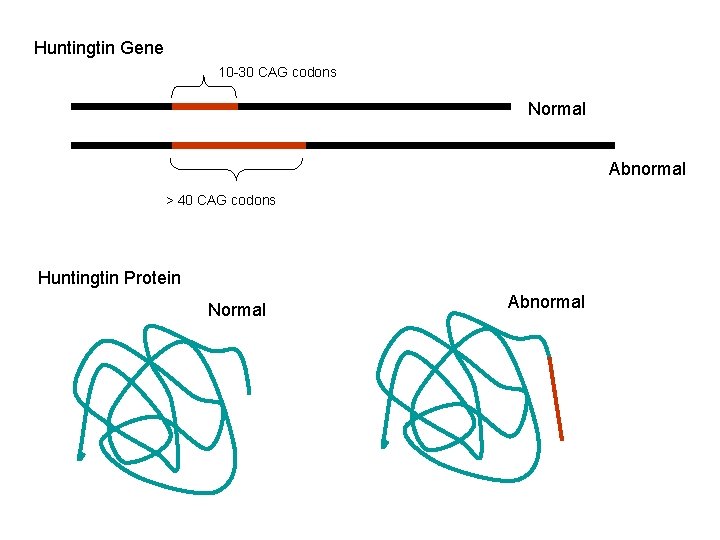
Huntington’s Disease Incidence 2 -8 persons per 100, 000 worldwide with focal population clusters Cause Known: excess of trinucleotide (CAG) repeats (encode glutamine) #CAG repeats 6 -34 Normal Gene 36 -120 HD Mutation (majority 40 -50 CAG repeats, 33 -40 yr onset) Number of repeats inversely related to age of onset. Juvenile onset is rare and involves CAG repeats >60.

Huntingtin Gene 10 -30 CAG codons Normal Abnormal > 40 CAG codons Huntingtin Protein Normal Abnormal

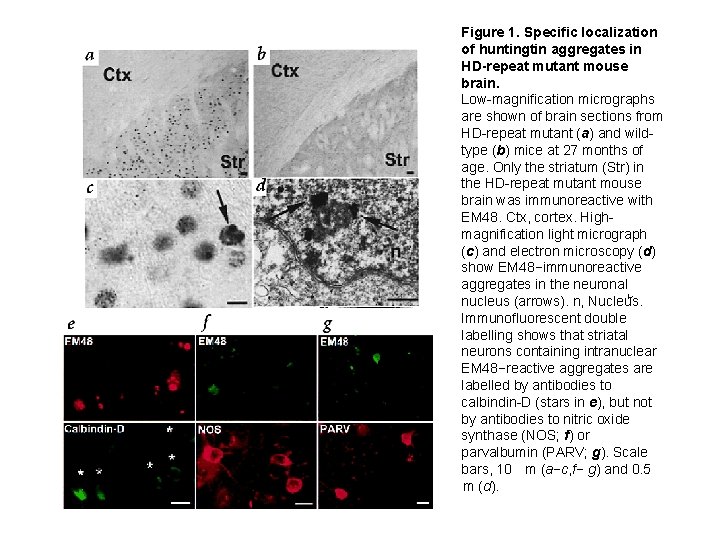
Figure 1. Specific localization of huntingtin aggregates in HD-repeat mutant mouse brain. Low-magnification micrographs are shown of brain sections from HD-repeat mutant (a) and wildtype (b) mice at 27 months of age. Only the striatum (Str) in the HD-repeat mutant mouse brain was immunoreactive with EM 48. Ctx, cortex. Highmagnification light micrograph (c) and electron microscopy (d) show EM 48−immunoreactive aggregates in the neuronal nucleus (arrows). n, Nucleus. Immunofluorescent double labelling shows that striatal neurons containing intranuclear EM 48−reactive aggregates are labelled by antibodies to calbindin-D (stars in e), but not by antibodies to nitric oxide synthase (NOS; f) or parvalbumin (PARV; g). Scale bars, 10 m (a−c, f− g) and 0. 5 m (d).

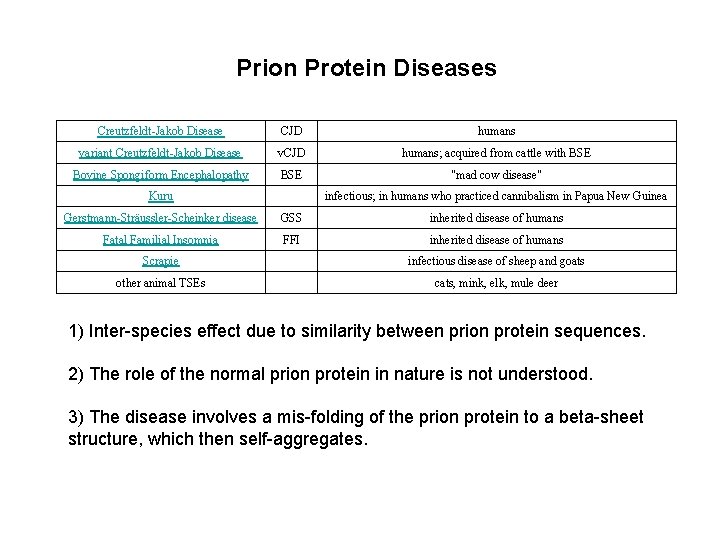
Prion Protein Diseases Creutzfeldt-Jakob Disease CJD humans variant Creutzfeldt-Jakob Disease v. CJD humans; acquired from cattle with BSE Bovine Spongiform Encephalopathy BSE "mad cow disease" Kuru infectious; in humans who practiced cannibalism in Papua New Guinea Gerstmann-Sträussler-Scheinker disease GSS inherited disease of humans Fatal Familial Insomnia FFI inherited disease of humans Scrapie infectious disease of sheep and goats other animal TSEs cats, mink, elk, mule deer 1) Inter-species effect due to similarity between prion protein sequences. 2) The role of the normal prion protein in nature is not understood. 3) The disease involves a mis-folding of the prion protein to a beta-sheet structure, which then self-aggregates.

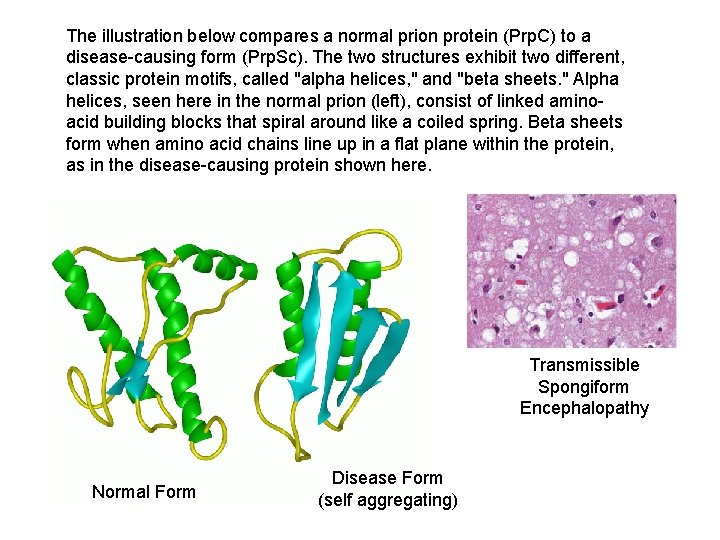
The illustration below compares a normal prion protein (Prp. C) to a disease-causing form (Prp. Sc). The two structures exhibit two different, classic protein motifs, called "alpha helices, " and "beta sheets. " Alpha helices, seen here in the normal prion (left), consist of linked aminoacid building blocks that spiral around like a coiled spring. Beta sheets form when amino acid chains line up in a flat plane within the protein, as in the disease-causing protein shown here. Transmissible Spongiform Encephalopathy Normal Form Disease Form (self aggregating)