Protein Structures 1 2 3 Fig 5 16




![Protein-ligand interaction n P+L Ka = PL [PL] [P] [L] Ka [L] = = Protein-ligand interaction n P+L Ka = PL [PL] [P] [L] Ka [L] = =](https://slidetodoc.com/presentation_image_h2/9ca44ab01ee5bba3b208b1976544d824/image-22.jpg)
![Ligand binding and Kd n n When [L] = Kd, 50% ligand-binding sites are Ligand binding and Kd n n When [L] = Kd, 50% ligand-binding sites are](https://slidetodoc.com/presentation_image_h2/9ca44ab01ee5bba3b208b1976544d824/image-23.jpg)
- Slides: 35

Protein Structures 1. 2. 3. Fig 5 -16 4. • • Primary structure Amino acid sequence Edman degradation, MS, deduce from DNA Secondary structure Recurring structural pattern Circular dichroism (CD, 圓二色極化光譜儀) Tertiary structure 3 D folding of a polypeptide chain X-ray crystallography, NMR Quaternary structure Subunits arrangement within a protein 1

The 3 -D structure of proteins • Protein conformation in space • Including long-range interactions • Determined by: • • • Primary (and secondary) structures Interactions among R groups Disulfide bond and weak interactions 2

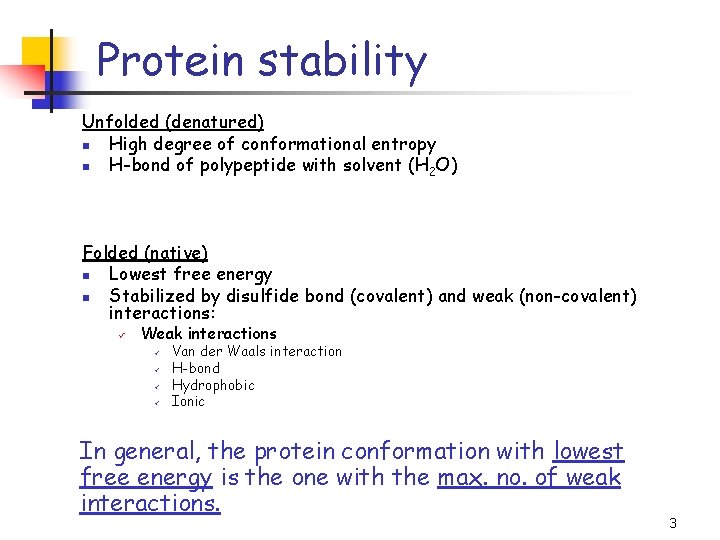
Protein stability Unfolded (denatured) n High degree of conformational entropy n H-bond of polypeptide with solvent (H 2 O) Folded (native) n Lowest free energy n Stabilized by disulfide bond (covalent) and weak (non-covalent) interactions: ü Weak interactions ü ü Van der Waals interaction H-bond Hydrophobic Ionic In general, the protein conformation with lowest free energy is the one with the max. no. of weak interactions. 3

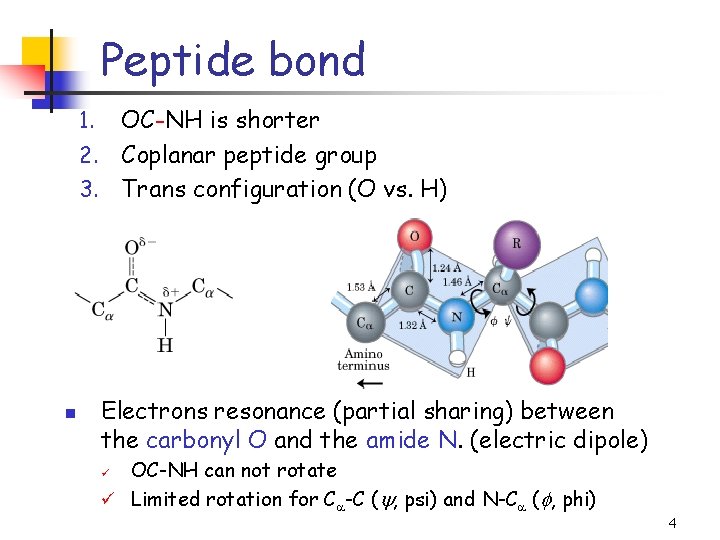
Peptide bond OC-NH is shorter 2. Coplanar peptide group 3. Trans configuration (O vs. H) 1. n Electrons resonance (partial sharing) between the carbonyl O and the amide N. (electric dipole) OC-NH can not rotate ü Limited rotation for Ca-C ( , psi) and N-Ca ( , phi) ü 4

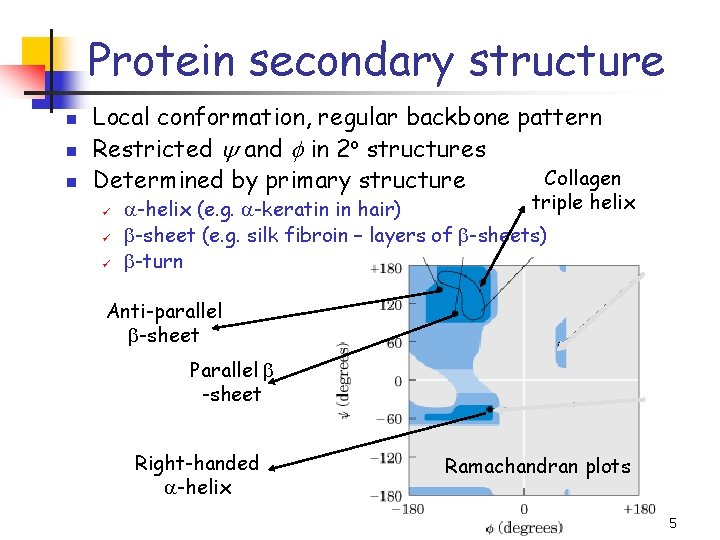
Protein secondary structure n n n Local conformation, regular backbone pattern Restricted and in 2 o structures Collagen Determined by primary structure ü ü ü triple helix a-helix (e. g. a-keratin in hair) b-sheet (e. g. silk fibroin – layers of b-sheets) b-turn Anti-parallel b-sheet Parallel b -sheet Right-handed a-helix Ramachandran plots 5

a-helix A right-handed a-helix: Ø Ø 3. 6 a. a. per turn 5. 4 Å (1 Å = 0. 1 nm) per turn R-R groups extended outward perpendicular to the helical axis H-bonding between adjacent turns Ø Ø 1 turn -- R H-bond between the -CO of residue (i) and the -NH of residue (i+3). 2 H-bonds per residue 3 or 4 H-bonds per turn Provide stability Box 6 -1 6

a-helix constraints 1. 2. 3. 4. 5. Electrostatic interactions of Ri and Ri+1 Size of the R group Interactions between Ri and Ri+3 or Ri+4 Pro and Gly End residues (electric dipole) O O H 3 N+ C CH H - O- H 3 N + CH 2 C OCH CH 2 + 7

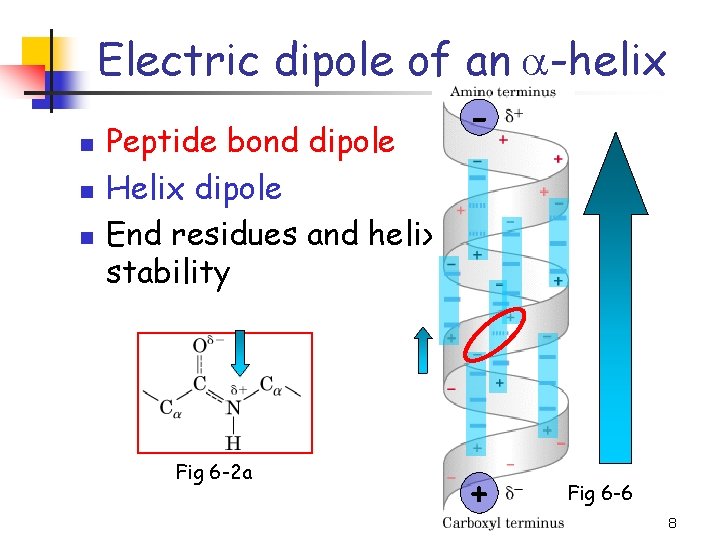
Electric dipole of an a-helix n n n Peptide bond dipole Helix dipole End residues and helix stability Fig 6 -2 a - + Fig 6 -6 8

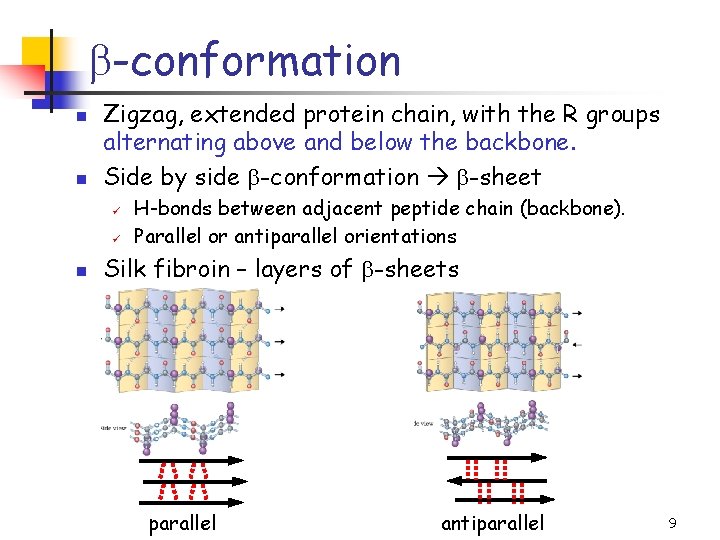
b-conformation n n Zigzag, extended protein chain, with the R groups alternating above and below the backbone. Side by side b-conformation b-sheet ü ü n H-bonds between adjacent peptide chain (backbone). Parallel or antiparallel orientations Silk fibroin – layers of b-sheets parallel antiparallel 9

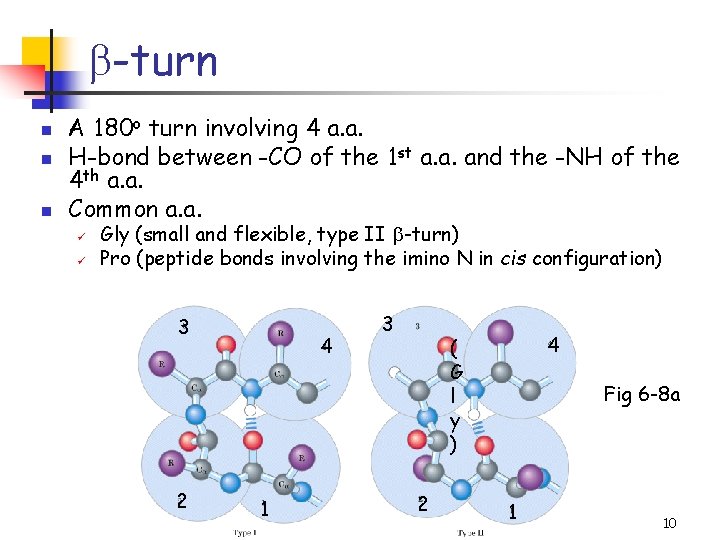
b-turn n A 180 o turn involving 4 a. a. H-bond between -CO of the 1 st a. a. and the -NH of the 4 th a. a. Common a. a. ü ü Gly (small and flexible, type II b-turn) Pro (peptide bonds involving the imino N in cis configuration) 3 2 4 1 3 4 ( G l y ) 2 Fig 6 -8 a 1 10

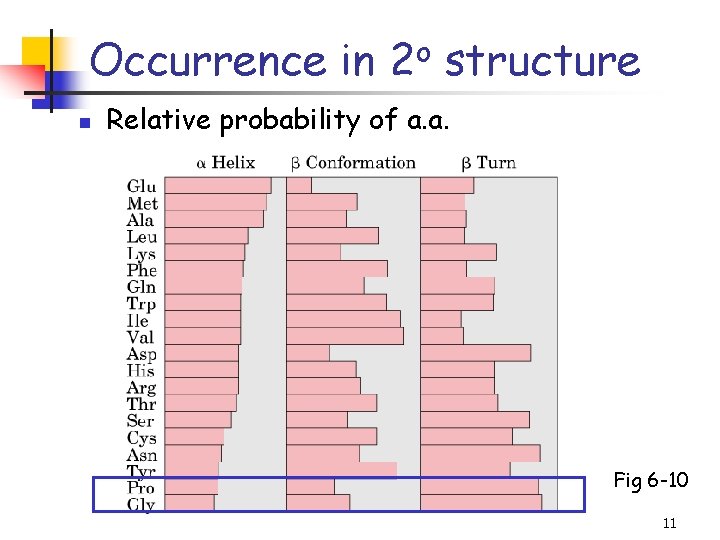
Occurrence in 2 o structure n Relative probability of a. a. Fig 6 -10 11

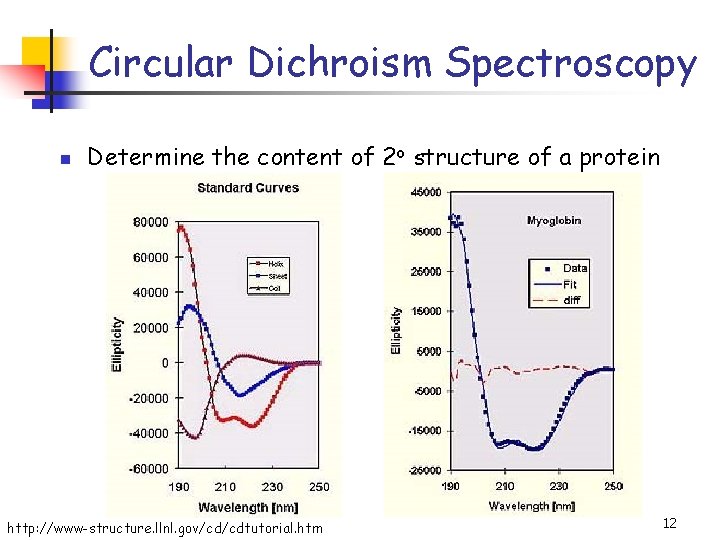
Circular Dichroism Spectroscopy n Determine the content of 2 o structure of a protein http: //www-structure. llnl. gov/cd/cdtutorial. htm 12

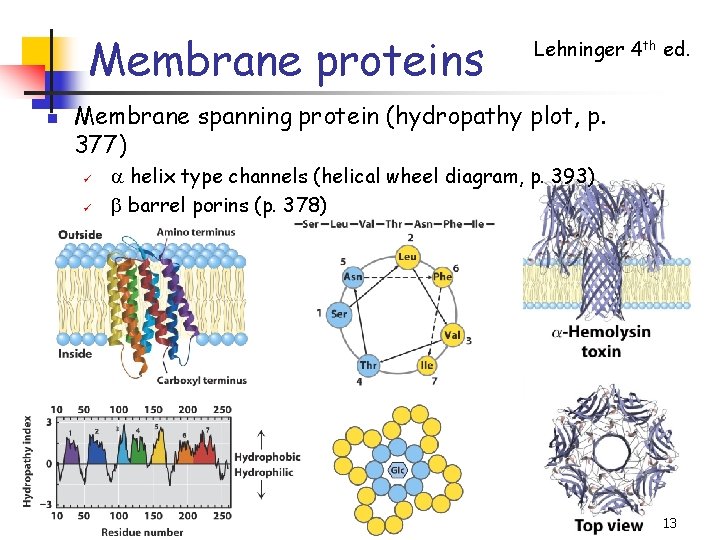
Membrane proteins n Lehninger 4 th ed. Membrane spanning protein (hydropathy plot, p. 377) ü ü a helix type channels (helical wheel diagram, p. 393) b barrel porins (p. 378) 13

Classification (p. 170) n Fibrous proteins (e. g. Table 6 -1) ü ü n Long strands or sheets Consist of a single type of 2 o structure Function in structure, support, protection a-keratin, collagen Globular proteins (e. g. Table 6 -2) ü ü Spherical or globular shape Contain several types of 2 o structure Function in regulation Myoglobin, hemoglobin 14

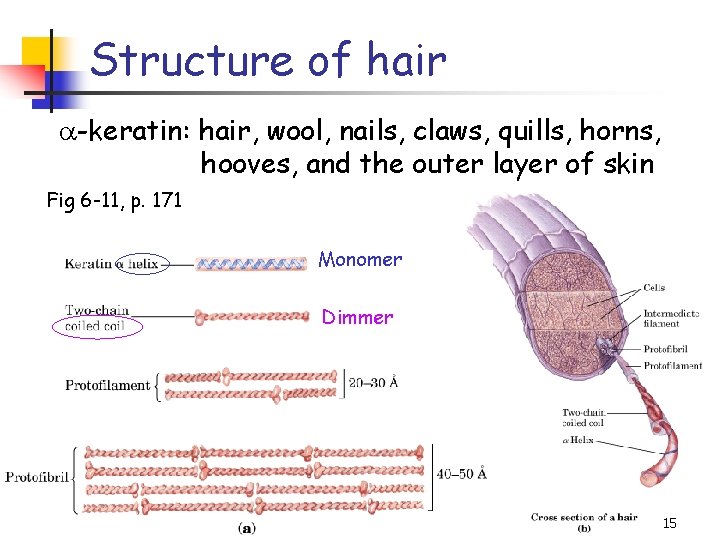
Structure of hair a-keratin: hair, wool, nails, claws, quills, horns, hooves, and the outer layer of skin Fig 6 -11, p. 171 Monomer Dimmer 15

Collagen n n Tendons, bone, cartilage, skin, and cornea Primary sequence: ü ü n Gly-X-Pro (Hy. Pro) Repeating tripeptide unit Structure ü Monomer (a chain) ü ü Left-handed helix, 3 a. a. per turn Trimer: coiled-coil (tensile strength). ü ü n collagen Stabilized by H-bond Crosslink between triple helixes tropocollagen Genetic defect: ü Osteogenesis imperfecta ü ü Abnormal bone formation in babies Ehlers-Danlos syndrome ü Loose joint 16

More on Collagen … Harper’s 26 th, p. 38 -39. n Procollagen (a larger precursor polypeptide) ü Post-translational modification ü ü Central portion triple helix (procollagen collagen) ü ü Pro, Lys Hydroxyl Pro, Lys (cofactor = ascorbic acid) Provide H-bond that stablizes the mature protein Scurvy: a dietary deficiency of Vit C The N-, and C-terminal portions are removed Certain Lys are modified by lysyl oxidase (a coppercontaining protein) ü ü Crosslink between polypeptides increased strength and rigidity. Menke’s syndrome: a dietary deficiency of the copper 17

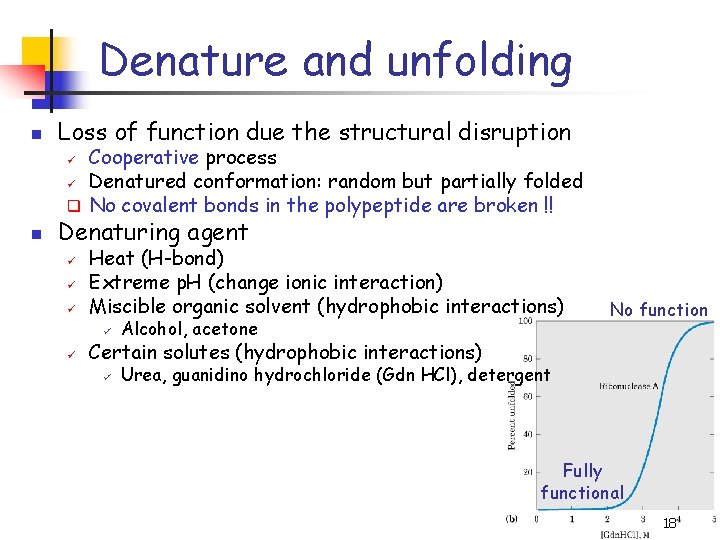
Denature and unfolding n Loss of function due the structural disruption Cooperative process ü Denatured conformation: random but partially folded q No covalent bonds in the polypeptide are broken !! ü n Denaturing agent ü ü Heat (H-bond) Extreme p. H (change ionic interaction) Miscible organic solvent (hydrophobic interactions) ü Alcohol, acetone ü Urea, guanidino hydrochloride (Gdn HCl), detergent No function Certain solutes (hydrophobic interactions) Fully functional 18

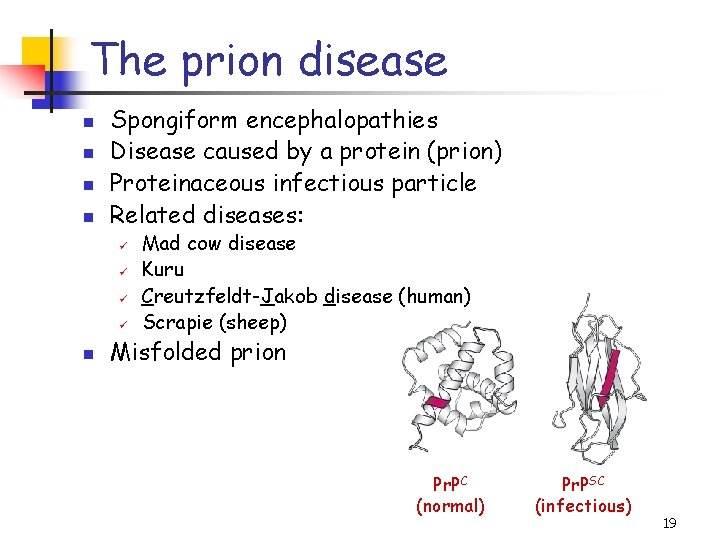
The prion disease n n Spongiform encephalopathies Disease caused by a protein (prion) Proteinaceous infectious particle Related diseases: ü ü n Mad cow disease Kuru Creutzfeldt-Jakob disease (human) Scrapie (sheep) Misfolded prion Pr. PC (normal) Pr. PSC (infectious) 19

Protein Function Myoglobin and Hemoglobin 20

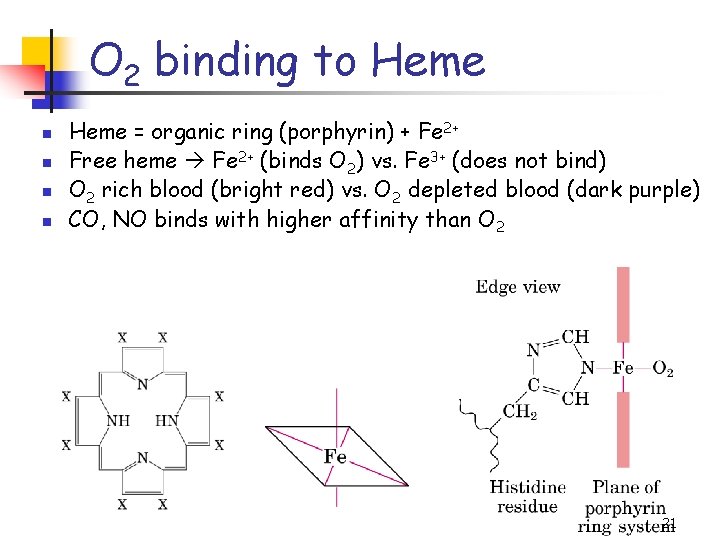
O 2 binding to Heme n n Heme = organic ring (porphyrin) + Fe 2+ Free heme Fe 2+ (binds O 2) vs. Fe 3+ (does not bind) O 2 rich blood (bright red) vs. O 2 depleted blood (dark purple) CO, NO binds with higher affinity than O 2 21
![Proteinligand interaction n PL Ka PL PL P L Ka L Protein-ligand interaction n P+L Ka = PL [PL] [P] [L] Ka [L] = =](https://slidetodoc.com/presentation_image_h2/9ca44ab01ee5bba3b208b1976544d824/image-22.jpg)
Protein-ligand interaction n P+L Ka = PL [PL] [P] [L] Ka [L] = = p. 207 Ka: association constant (M-1) [PL] [P] Binding sites occupied Total binding sites [L] = = [L] + 1/Ka [L] + Kd [PL] = [PL] + [P] Kd: dissociation constant (M) 22
![Ligand binding and Kd n n When L Kd 50 ligandbinding sites are Ligand binding and Kd n n When [L] = Kd, 50% ligand-binding sites are](https://slidetodoc.com/presentation_image_h2/9ca44ab01ee5bba3b208b1976544d824/image-23.jpg)
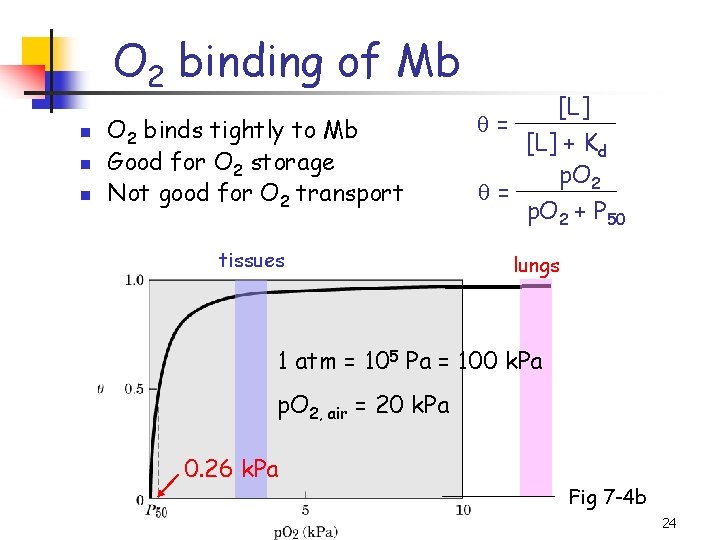
Ligand binding and Kd n n When [L] = Kd, 50% ligand-binding sites are occupied Kd: dissociation constant Kd = [L] at half-saturation Affinity , Kd Hyperbola [L] = [L] + Kd Fig 7 -4 a 23

O 2 binding of Mb n n n O 2 binds tightly to Mb Good for O 2 storage Not good for O 2 transport tissues [L] = [L] + Kd p. O 2 = p. O 2 + P 50 lungs 1 atm = 105 Pa = 100 k. Pa p. O 2, air = 20 k. Pa 0. 26 k. Pa Fig 7 -4 b 24

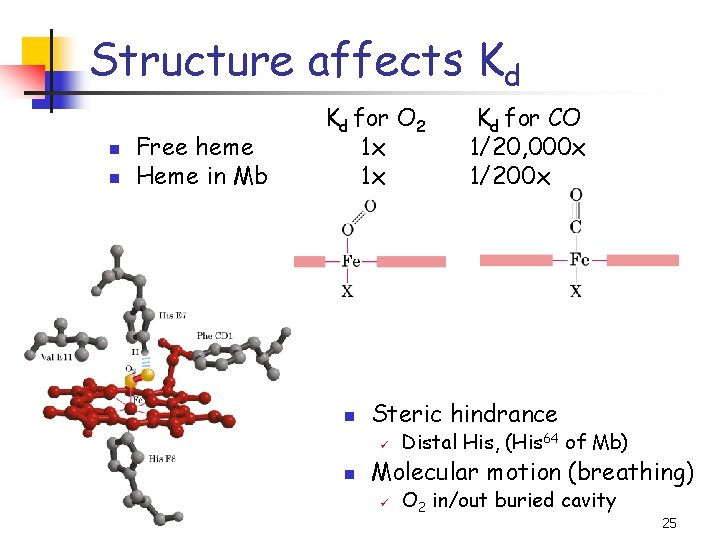
Structure affects Kd n n Free heme Heme in Mb Kd for O 2 1 x 1 x n Steric hindrance ü n Kd for CO 1/20, 000 x 1/200 x Distal His, (His 64 of Mb) Molecular motion (breathing) ü O 2 in/out buried cavity 25

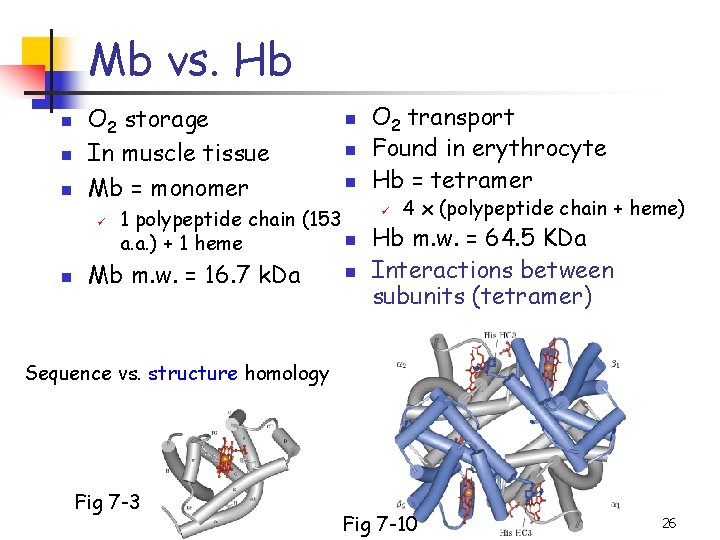
Mb vs. Hb n n n O 2 storage In muscle tissue Mb = monomer ü n n 1 polypeptide chain (153 n a. a. ) + 1 heme Mb m. w. = 16. 7 k. Da n O 2 transport Found in erythrocyte Hb = tetramer ü 4 x (polypeptide chain + heme) Hb m. w. = 64. 5 KDa Interactions between subunits (tetramer) Sequence vs. structure homology Fig 7 -3 Fig 7 -10 26

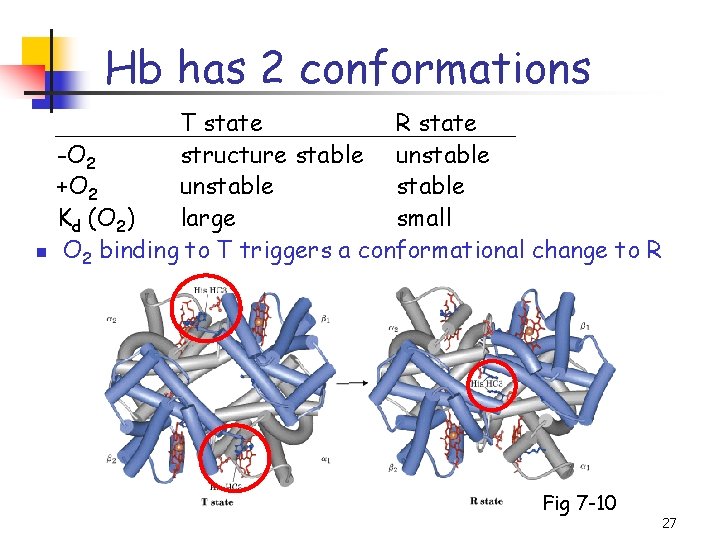
Hb has 2 conformations n T state R state -O 2 structure stable unstable +O 2 unstable Kd (O 2) large small O 2 binding to T triggers a conformational change to R Fig 7 -10 27

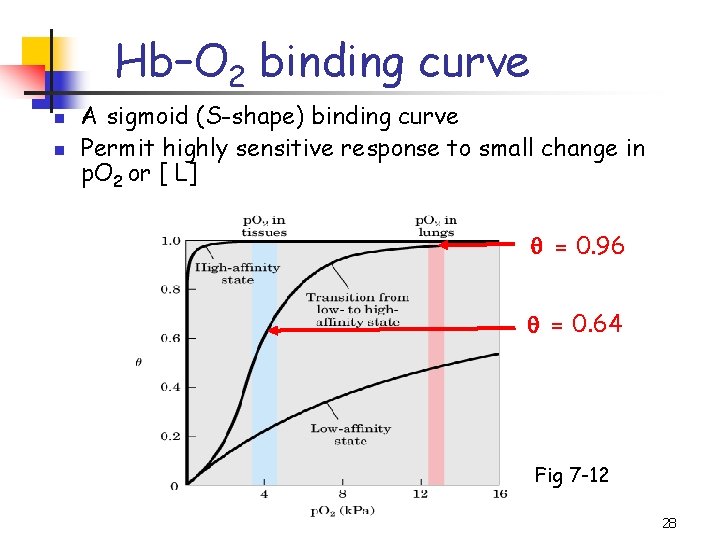
Hb–O 2 binding curve n n A sigmoid (S-shape) binding curve Permit highly sensitive response to small change in p. O 2 or [ L] = 0. 96 = 0. 64 Fig 7 -12 28

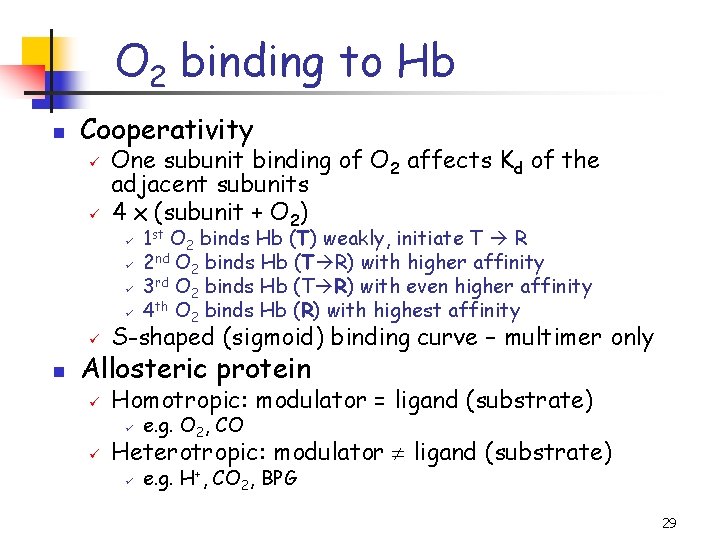
O 2 binding to Hb n Cooperativity ü ü One subunit binding of O 2 affects Kd of the adjacent subunits 4 x (subunit + O 2) ü ü n 1 st O 2 binds Hb (T) weakly, initiate T R 2 nd O 2 binds Hb (T R) with higher affinity 3 rd O 2 binds Hb (T R) with even higher affinity 4 th O 2 binds Hb (R) with highest affinity ü S-shaped (sigmoid) binding curve – multimer only ü Homotropic: modulator = ligand (substrate) Allosteric protein ü ü e. g. O 2, CO ü e. g. H+, CO 2, BPG Heterotropic: modulator ligand (substrate) 29

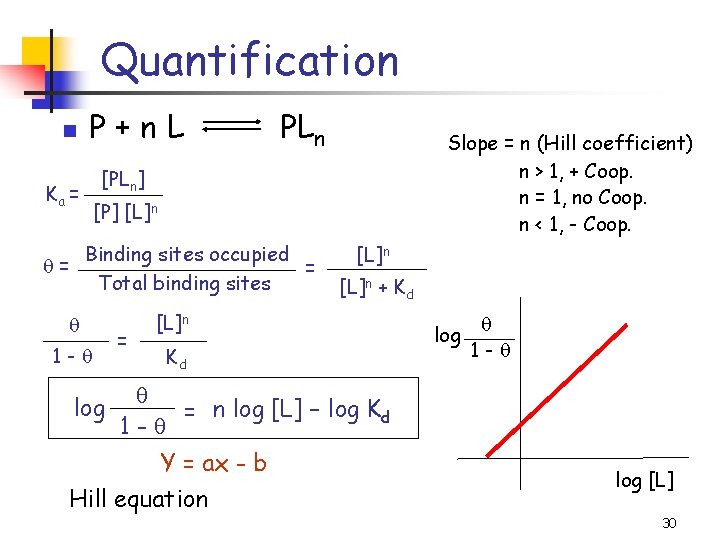
Quantification n P+n. L Ka = PLn Slope = n (Hill coefficient) n > 1, + Coop. n = 1, no Coop. n < 1, - Coop. [PLn] [P] [L]n Binding sites occupied = = Total binding sites 1 - log = [L]n + Kd [L]n Kd log 1 - = n log [L] – log Kd 1 - Y = ax - b Hill equation log [L] 30

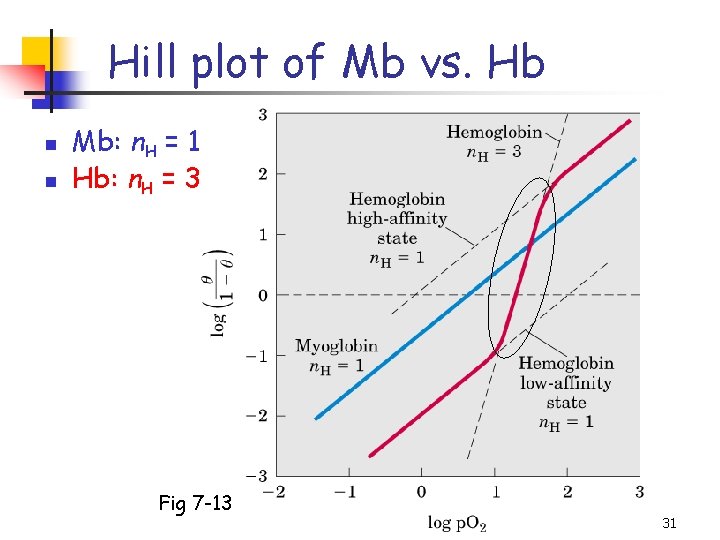
Hill plot of Mb vs. Hb n n Mb: n. H = 1 Hb: n. H = 3 Fig 7 -13 31

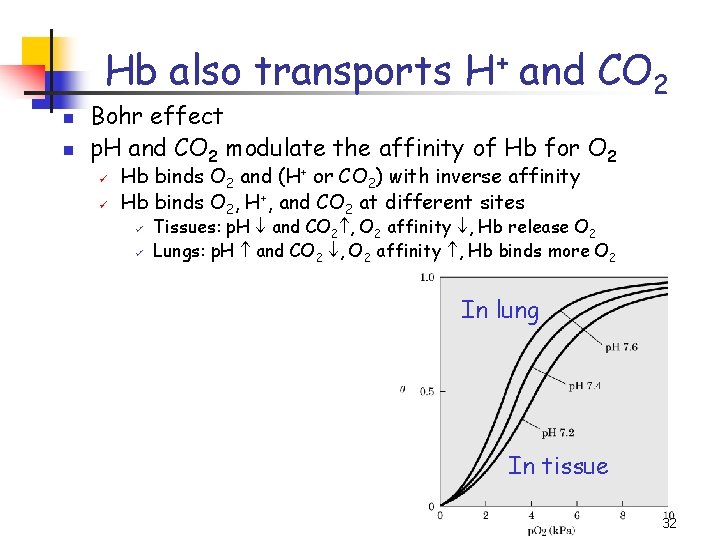
Hb also transports H+ and CO 2 n n Bohr effect p. H and CO 2 modulate the affinity of Hb for O 2 ü ü Hb binds O 2 and (H+ or CO 2) with inverse affinity Hb binds O 2, H+, and CO 2 at different sites ü ü Tissues: p. H and CO 2 , O 2 affinity , Hb release O 2 Lungs: p. H and CO 2 , O 2 affinity , Hb binds more O 2 In lung In tissue 32

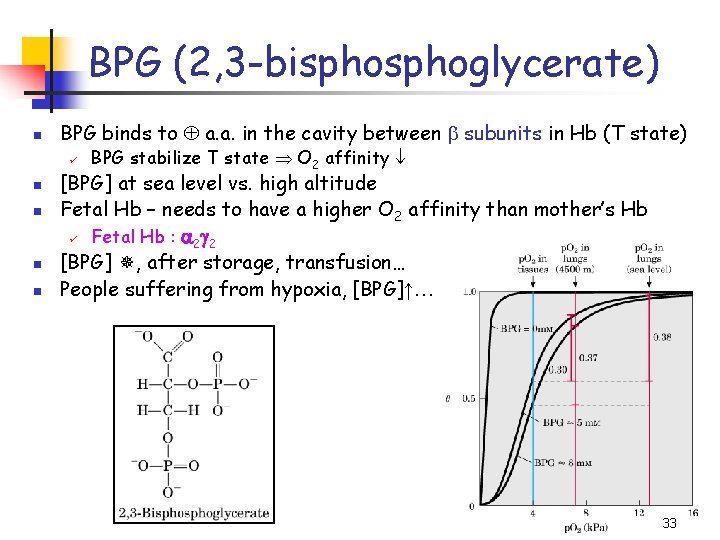
BPG (2, 3 -bisphoglycerate) n BPG binds to a. a. in the cavity between b subunits in Hb (T state) ü n n BPG stabilize T state O 2 affinity [BPG] at sea level vs. high altitude Fetal Hb – needs to have a higher O 2 affinity than mother’s Hb ü Fetal Hb : a 2 g 2 [BPG] , after storage, transfusion… People suffering from hypoxia, [BPG]↑… 33

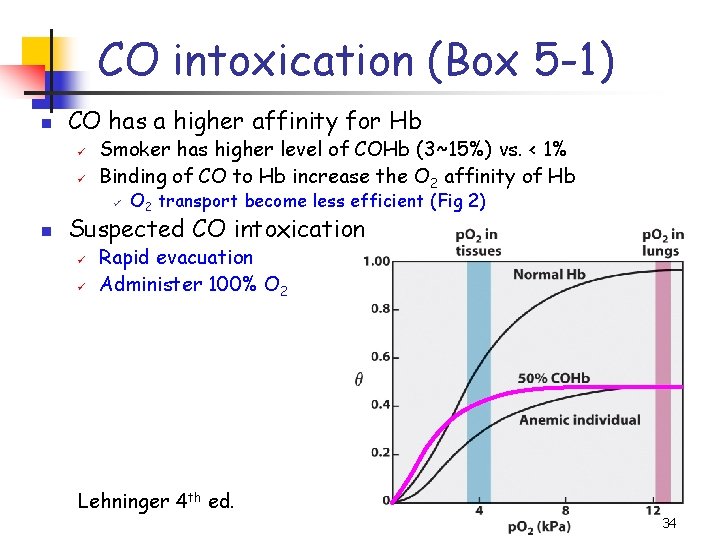
CO intoxication (Box 5 -1) n CO has a higher affinity for Hb ü ü Smoker has higher level of COHb (3~15%) vs. < 1% Binding of CO to Hb increase the O 2 affinity of Hb ü n O 2 transport become less efficient (Fig 2) Suspected CO intoxication ü ü Rapid evacuation Administer 100% O 2 Lehninger 4 th ed. 34

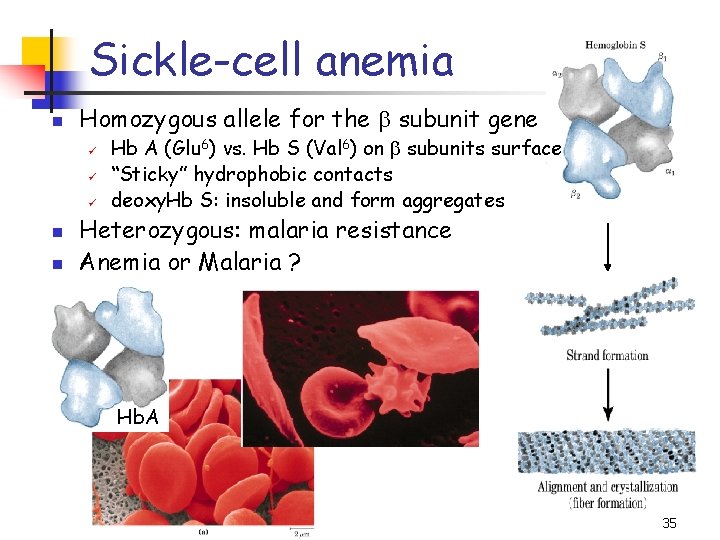
Sickle-cell anemia n Homozygous allele for the b subunit gene ü ü ü n n Hb A (Glu 6) vs. Hb S (Val 6) on b subunits surface “Sticky” hydrophobic contacts deoxy. Hb S: insoluble and form aggregates Heterozygous: malaria resistance Anemia or Malaria ? Hb. A 35